Volume 12, Issue 2 (March & April 2021)
BCN 2021, 12(2): 187-198 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Zamanian-Azodi M, Rezaei-Tavirani M, Rezaei Tavirani M. Obsessive-Compulsive Disorder Interactome Profile Analysis: A Perspective From Molecular Mechanism. BCN 2021; 12 (2) :187-198
URL: http://bcn.iums.ac.ir/article-1-1590-en.html
URL: http://bcn.iums.ac.ir/article-1-1590-en.html
1- Proteomics Research Center, Faculty of Paramedical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Keywords: Obsessive-Compulsive Disorder (OCD), Protein-Protein Interaction (PPI) network, Gene Ontology annotation (GO)
Full-Text [PDF 1035 kb]
| Abstract (HTML)
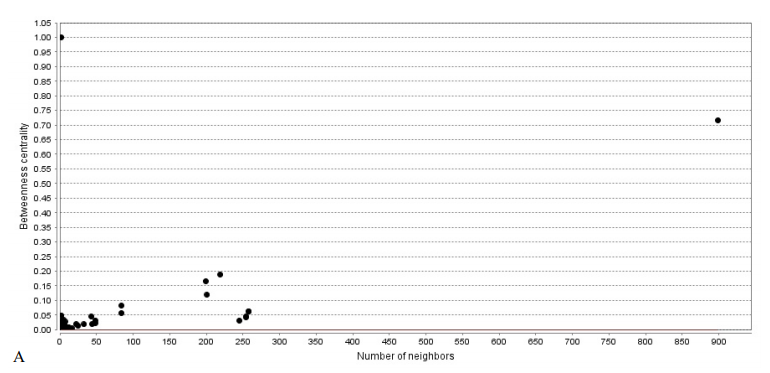
The PPI network of this study is formed by 1940 nodes and connected via 3269 edges. Graph topological properties help gain insight into the global network structure (Assenov et al., 2008). Node distribution based on centrality parameters such as K and BC shows the presence of genes with high centrality values (Figure 1).
The top ten central genes are presented in Table 2. The higher the value, the more central the nodes are in the network (Scardoni, Petterlini, & Laudanna, 2009).
In Figure 1, K values are significantly heterogeneous. Just a few nodes show well-linked, whereas others possess a small number of connections. This distribution implies the presents of genes with high centrality values. The line indicates the power law. The R2 value is computed on logarithmic values, which is equal to 0.53, and the correlation is 0.84. Genes with a high degree are in the right down region of the plot (out of linear range). B: R=0.002 and correlation=0.38, The BT range is from 0-1. Nodes in the top and middle regions of the plot (out of linear range) have high BC values.
The amounts of degree and betweenness centralities for the introduced key genes are represented in Table 2. Based on Table 2, degree values for the hub-bottleneck nodes (except ESR1) vary between 200 and 250.
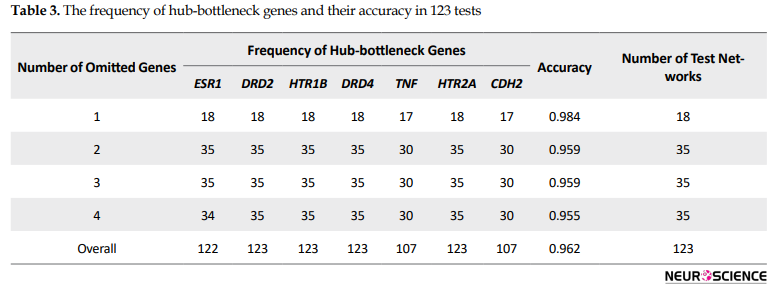
A backbone network of the introduced 7 hub-bottleneck nodes with the related topological features was obtained (the data are not shown). Furthermore, to confirm the backbone network, we used the cross-validation method. Accordingly, 123 test networks (18+3×35) were constructed by deleting 1 to 4 nodes from 18 top nodes (largest degree). At first, the deletion of one out of 18 top nodes for each test resulted in the construction of 18 networks. Then, as deleting more than 2 genes would result in many different combinations, the test was done .
five times by selecting one hub-bottleneck gene and randomly selecting from other top genes (Table 3, Figure 2).
A backbone network was built from Mentha, Reactome, and Reactome-Fls sources for 7 hub-bottleneck elements. This network has 1609 nodes, and 2279 edges denote that 83% of the nodes relative to the main network are presented in the backbone network.
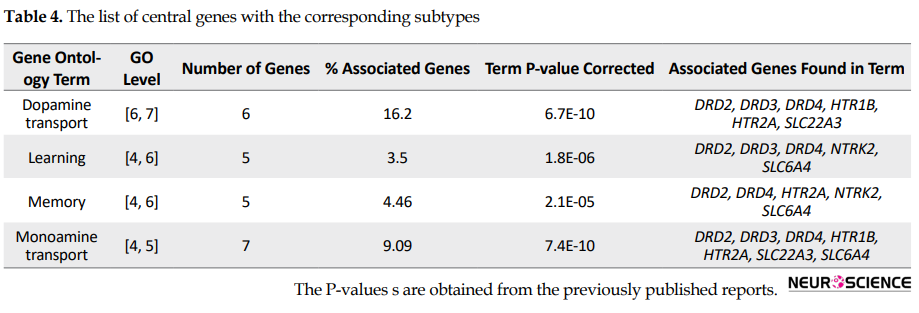
Different subtypes have been reported for OCD that may appear due to the interaction of different genes and environmental factors. Central genes in this study are reported with different subtypes (Table 4).
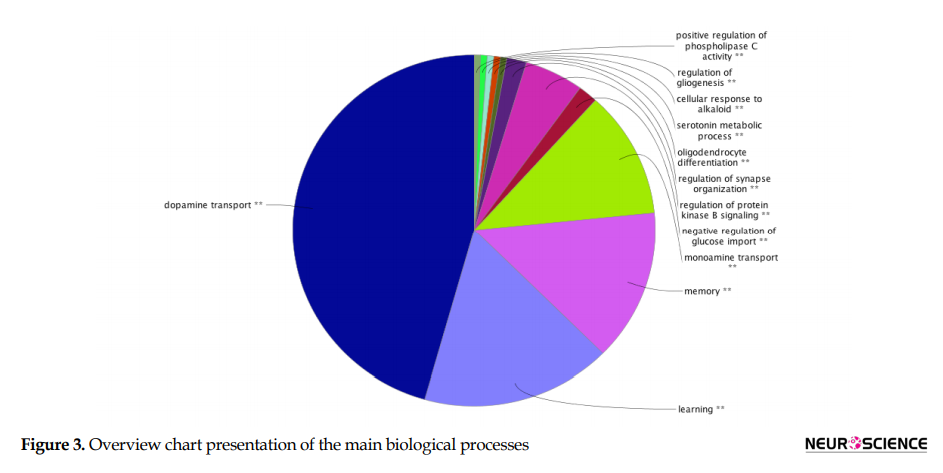
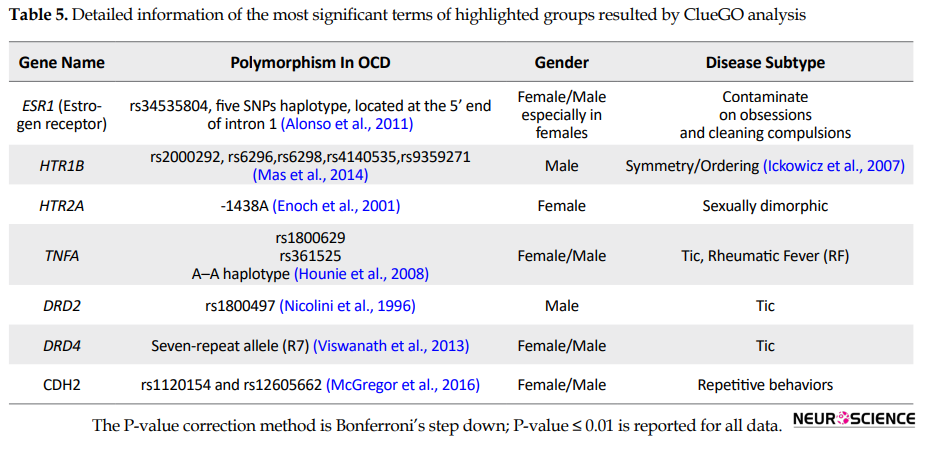
Application of ClueGO plugin for functional enrichment analysis of OCD seed genes based on BP terms is shown in Figure 3 and Table 5.
Full-Text:
1. Introduction
Obsessive-Compulsive Disorder (OCD) is known to be the fourth most frequent heterogeneous mental condition in the world. The lifetime prevalence of OCD is about 1% to 3% (Tang et al., 2014), and it is typically featured with unwanted thoughts and compulsive behavior (Zamanian-Azodi et al., 2015). The complex nature of OCD is correlated to the interactions of many genetic and environmental factors (Taylor, Asmundson, & Jang, 2016). These interactions can lead to complex clinical demonstration and symptom overlap of OCD with other related disorders, including Tourette syndrome, chronic hair pulling, trichotillomania, and anxiety, hindering proper therapeutic approaches (Mahboubi et al., 2018). Many genes with their polymorphisms have been introduced for OCD etiology (Taylor, 2013). However, the contribution of each gene is known to be partial (Jaffe et al., 2014). This fact implies the importance of examining genetic elements in a whole interaction system rather than singly (Rezaei-Tavirani, Tavirani, & Azodi, 2019).Any minor changes in these elements may influence the molecular interaction pattern and biological processes necessary for everyday functions. These alterations can promote the onset and development of a disease (Chavez, Weisbrod, Zheng, Eng, & Bruce, 2013). In this regard, protein-protein interaction network analysis can help map the interactome profile of a disease (Zamanian-Azodi et al., 2018). Identifying central genes in a predicted network is valuable for better understanding potential elements of disease underlying mechanisms and accordingly introducing possible therapeutic targets (Mahboubi et al., 2018). As mentioned earlier, many genes are reported to be associated with OCD risk. However, there are inconsistencies in their relations among studies. This might be due to various factors, including the difference in sample size, subtypes, Studied single Nucleotide Polymorphisms (SNPs), and the applied statistical criteria. In this regard, a comprehensive meta-analysis may provide more information. Based on one meta-analysis, there are significant relationships between OCD and some genes from genetic association studies (Taylor, 2013).
Nevertheless, this approach may not be adequate for network analysis since some of the other genes that were clinically showed association with OCD might be missed. For instance, dopaminergic system genes showed a lack of significant associations with OCD in the mentioned meta-analysis, while treatment with dopamine receptor antagonism proved to be effective (Koo, Kim, Roh, & Kim, 2010; Lochner et al., 2016). The heterogeneous basis of the genetic association studies made it difficult for the PPI network dataset design. At least 31 genes have been introduced for OCD onset and development, but there is not an explicit priority between them. Therefore, determination of the crucial genes in OCD by interactome analysis is the aim of this investigation.
2. Methods
Data visualization in the form of complex networks can be obtained by applying Cytoscape (an open-source network analyzer tool; http://cytoscape.org) (Saito et al., 2012). For analyzing genes from genetic association studies, 31 genes for OCD patients were searched through popular literature motor engines (Google Scholar) and specialized biomedical databank (PubMed). The first step for network analysis was to retrieve UniProt accession codes (Homo sapiens) of the 31 genes from http://www.uniprot.org. Then the codes were used for PPI mapping by Cytoscape v: 3.2.1. For PPI network construction, PSICQUIC (Proteomics Standard Initiative Common QUery InterfaCe) was applied. This algorithm is a universal web service client for importing interactions from public databases (Aranda et al., 2011). The applied public databases in this study were Mentha, Reactome-Fls, and Reactome (Lotia, Montojo, Dong, Bader, & Pico, 2013).
Further analysis was performed based on functional topological parameters (centrality), including node degree and betweenness centrality, using Network Analyzer Application. This software is a versatile and user-friendly tool for the analysis of biological networks. This plugin is well-integrated into Cytoscape and computes a comprehensive list of complex and straightforward topology parameters using resourceful graphic algorithms (Assenov, Ramírez, Schelhorn, Lengauer, & Albrecht, 2008). In the PPI network, the nodes represent genes, while the edges represent physical interactions (Luo et al., 2014). Degree (K) implies the number of connected edges to a consider vertices. Betweenness Centrality (BC) equals the number of shortest paths that pass through each node (Assenov et al., 2008). Genes with large K and high BC are called hub-bottlenecks. For hub-bottleneck validation, test networks were obtained based on omitting (1 to 4) top nodes of the main network. It should be noted that for deleting more than one node, a combination of one hub-bottleneck gene and random selection from the remained top genes with five times repetition was performed. In the end, the robustness of the backbone network was calculated (Nair, Ghatge, Kakkar, & Shanker, 2014). ClueGO plugin is an up-to-date functional analyzer that enriches a list of genes based on gene ontology (Bindea et al., 2009). Here, GlueGO2.1.7 annotates genes for biological process. ClueGO presents gene ontology ranging from general to specific ones. Biological terms were assessed for the designated 31 OCD genes. Genes were grouped based on a significant functional association between terms and gene-sets. For redundancy reduction, the fusion of the relevant terms with the related genes was applied. The association strength between the terms is calculated by Kappa scoring. The Kappa score level was adjusted to 0.5 (medium) for Biological Process (BP) analysis. In this analysis, the minimum number and percent. age of genes per term were adjusted to 3 and 4 as the default options, respectively. Likewise, minimum and maximum levels of ontology were set to 3 and 8 as the default option, respectively. The P value was also set to ≤0.05, which was corrected by the Bonferroni step-down method. The enrichment/depletion test for terms was set to two-sided (enrichment/depletion) based on hypergeometric (Bindea, Galon, & Mlecnik, 2013).
3. Results
Thirty-one OCD seed genes with their properties were retrieved from the genetic association studies as tabulated in Table 1.
Obsessive-Compulsive Disorder (OCD) is known to be the fourth most frequent heterogeneous mental condition in the world. The lifetime prevalence of OCD is about 1% to 3% (Tang et al., 2014), and it is typically featured with unwanted thoughts and compulsive behavior (Zamanian-Azodi et al., 2015). The complex nature of OCD is correlated to the interactions of many genetic and environmental factors (Taylor, Asmundson, & Jang, 2016). These interactions can lead to complex clinical demonstration and symptom overlap of OCD with other related disorders, including Tourette syndrome, chronic hair pulling, trichotillomania, and anxiety, hindering proper therapeutic approaches (Mahboubi et al., 2018). Many genes with their polymorphisms have been introduced for OCD etiology (Taylor, 2013). However, the contribution of each gene is known to be partial (Jaffe et al., 2014). This fact implies the importance of examining genetic elements in a whole interaction system rather than singly (Rezaei-Tavirani, Tavirani, & Azodi, 2019).Any minor changes in these elements may influence the molecular interaction pattern and biological processes necessary for everyday functions. These alterations can promote the onset and development of a disease (Chavez, Weisbrod, Zheng, Eng, & Bruce, 2013). In this regard, protein-protein interaction network analysis can help map the interactome profile of a disease (Zamanian-Azodi et al., 2018). Identifying central genes in a predicted network is valuable for better understanding potential elements of disease underlying mechanisms and accordingly introducing possible therapeutic targets (Mahboubi et al., 2018). As mentioned earlier, many genes are reported to be associated with OCD risk. However, there are inconsistencies in their relations among studies. This might be due to various factors, including the difference in sample size, subtypes, Studied single Nucleotide Polymorphisms (SNPs), and the applied statistical criteria. In this regard, a comprehensive meta-analysis may provide more information. Based on one meta-analysis, there are significant relationships between OCD and some genes from genetic association studies (Taylor, 2013).
Nevertheless, this approach may not be adequate for network analysis since some of the other genes that were clinically showed association with OCD might be missed. For instance, dopaminergic system genes showed a lack of significant associations with OCD in the mentioned meta-analysis, while treatment with dopamine receptor antagonism proved to be effective (Koo, Kim, Roh, & Kim, 2010; Lochner et al., 2016). The heterogeneous basis of the genetic association studies made it difficult for the PPI network dataset design. At least 31 genes have been introduced for OCD onset and development, but there is not an explicit priority between them. Therefore, determination of the crucial genes in OCD by interactome analysis is the aim of this investigation.
2. Methods
Data visualization in the form of complex networks can be obtained by applying Cytoscape (an open-source network analyzer tool; http://cytoscape.org) (Saito et al., 2012). For analyzing genes from genetic association studies, 31 genes for OCD patients were searched through popular literature motor engines (Google Scholar) and specialized biomedical databank (PubMed). The first step for network analysis was to retrieve UniProt accession codes (Homo sapiens) of the 31 genes from http://www.uniprot.org. Then the codes were used for PPI mapping by Cytoscape v: 3.2.1. For PPI network construction, PSICQUIC (Proteomics Standard Initiative Common QUery InterfaCe) was applied. This algorithm is a universal web service client for importing interactions from public databases (Aranda et al., 2011). The applied public databases in this study were Mentha, Reactome-Fls, and Reactome (Lotia, Montojo, Dong, Bader, & Pico, 2013).
Further analysis was performed based on functional topological parameters (centrality), including node degree and betweenness centrality, using Network Analyzer Application. This software is a versatile and user-friendly tool for the analysis of biological networks. This plugin is well-integrated into Cytoscape and computes a comprehensive list of complex and straightforward topology parameters using resourceful graphic algorithms (Assenov, Ramírez, Schelhorn, Lengauer, & Albrecht, 2008). In the PPI network, the nodes represent genes, while the edges represent physical interactions (Luo et al., 2014). Degree (K) implies the number of connected edges to a consider vertices. Betweenness Centrality (BC) equals the number of shortest paths that pass through each node (Assenov et al., 2008). Genes with large K and high BC are called hub-bottlenecks. For hub-bottleneck validation, test networks were obtained based on omitting (1 to 4) top nodes of the main network. It should be noted that for deleting more than one node, a combination of one hub-bottleneck gene and random selection from the remained top genes with five times repetition was performed. In the end, the robustness of the backbone network was calculated (Nair, Ghatge, Kakkar, & Shanker, 2014). ClueGO plugin is an up-to-date functional analyzer that enriches a list of genes based on gene ontology (Bindea et al., 2009). Here, GlueGO2.1.7 annotates genes for biological process. ClueGO presents gene ontology ranging from general to specific ones. Biological terms were assessed for the designated 31 OCD genes. Genes were grouped based on a significant functional association between terms and gene-sets. For redundancy reduction, the fusion of the relevant terms with the related genes was applied. The association strength between the terms is calculated by Kappa scoring. The Kappa score level was adjusted to 0.5 (medium) for Biological Process (BP) analysis. In this analysis, the minimum number and percent. age of genes per term were adjusted to 3 and 4 as the default options, respectively. Likewise, minimum and maximum levels of ontology were set to 3 and 8 as the default option, respectively. The P value was also set to ≤0.05, which was corrected by the Bonferroni step-down method. The enrichment/depletion test for terms was set to two-sided (enrichment/depletion) based on hypergeometric (Bindea, Galon, & Mlecnik, 2013).
3. Results
Thirty-one OCD seed genes with their properties were retrieved from the genetic association studies as tabulated in Table 1.
The PPI network of this study is formed by 1940 nodes and connected via 3269 edges. Graph topological properties help gain insight into the global network structure (Assenov et al., 2008). Node distribution based on centrality parameters such as K and BC shows the presence of genes with high centrality values (Figure 1).
The top ten central genes are presented in Table 2. The higher the value, the more central the nodes are in the network (Scardoni, Petterlini, & Laudanna, 2009).
In Figure 1, K values are significantly heterogeneous. Just a few nodes show well-linked, whereas others possess a small number of connections. This distribution implies the presents of genes with high centrality values. The line indicates the power law. The R2 value is computed on logarithmic values, which is equal to 0.53, and the correlation is 0.84. Genes with a high degree are in the right down region of the plot (out of linear range). B: R=0.002 and correlation=0.38, The BT range is from 0-1. Nodes in the top and middle regions of the plot (out of linear range) have high BC values.
The amounts of degree and betweenness centralities for the introduced key genes are represented in Table 2. Based on Table 2, degree values for the hub-bottleneck nodes (except ESR1) vary between 200 and 250.
A backbone network of the introduced 7 hub-bottleneck nodes with the related topological features was obtained (the data are not shown). Furthermore, to confirm the backbone network, we used the cross-validation method. Accordingly, 123 test networks (18+3×35) were constructed by deleting 1 to 4 nodes from 18 top nodes (largest degree). At first, the deletion of one out of 18 top nodes for each test resulted in the construction of 18 networks. Then, as deleting more than 2 genes would result in many different combinations, the test was done .
five times by selecting one hub-bottleneck gene and randomly selecting from other top genes (Table 3, Figure 2).
A backbone network was built from Mentha, Reactome, and Reactome-Fls sources for 7 hub-bottleneck elements. This network has 1609 nodes, and 2279 edges denote that 83% of the nodes relative to the main network are presented in the backbone network.
Different subtypes have been reported for OCD that may appear due to the interaction of different genes and environmental factors. Central genes in this study are reported with different subtypes (Table 4).
Application of ClueGO plugin for functional enrichment analysis of OCD seed genes based on BP terms is shown in Figure 3 and Table 5.
Groups are labeled based on the leading term (the most significant). The most significant term is defined as the term with the highest number and percentage of initial genes. Kappa Score ≥0.5. Figure 3 showed that dopamine transport, learning, memory, and monoamine transport are the main BPs.
five times by selecting one hub-bottleneck gene and randomly selecting from other top genes (Table 3, Figure 2).
A backbone network was built from Mentha, Reactome, and Reactome-Fls sources for 7 hub-bottleneck elements. This network has 1609 nodes, and 2279 edges denote that 83% of the nodes relative to the main network are presented in the backbone network.
Different subtypes have been reported for OCD that may appear due to the interaction of different genes and environmental factors. Central genes in this study are reported with different subtypes (Table 4).
Application of ClueGO plugin for functional enrichment analysis of OCD seed genes based on BP terms is shown in Figure 3 and Table 5.
Groups are labeled based on the leading term (the most significant). The most significant term is defined as the term with the highest number and percentage of initial genes. Kappa Score ≥0.5. Figure 3 showed that dopamine transport, learning, memory, and monoamine transport are the main BPs.
4. Discussion
Molecular studies of Obsessive-Compulsive Disorder (OCD) have just begun (Zamanian-Azodi et al., 2015). Most of the recent investigations are focused on the genetic basis of OCD. Multiple genetic variants with small effect size are linked to OCD risk (Gruenblatt, Hauser, & Walitza, 2014). While individual examining of genes is informative, interactome evaluation is essential as well. Genes are in a complex interaction structure that any genetic alterations may lead to functional phenotype changes of the whole system (Chavez et al., 2013). PPI network analysis can identify specific interactome changes in a disease state. In this regard, examining the OCD interactome pattern can help gain further insight into the underlying mechanisms through topological analysis (Mahboubi et al., 2018). In this study, a total number of 31 human candidate genes from genetic association studies are reviewed. Among them, 8 genes were previously reported significant for OCD by a meta-analysis (Taylor, 2013).
On the other hand, none of the single candidate genes has been proved to be significantly associated with OCD by genome-wide association studies (Gruenblatt et al., 2014). Moreover, mRNA expression profiling of postmortem brain of OCD patients showed no significant associations of these genes and mRNA expression profile (Jaffe et al., 2014). Therefore, a network from reported genes may be inaccurate since they are incomplete and may show some bias towards neurotransmitter systems genes (Gruenblatt et al., 2014). To somehow overcome this limitation, it is tried to select genes from genetic-based studies that were with at least one significant association report with OCD.
There are about 1.5 edges per node; however, this value’s variation is considerable for different nodes. For example, ESR1, the top node, is characterized by 899 links. This gene is discriminated by about 650 links from the second top score node. In a PPI network, central genes (hub-bottlenecks) are essential for network integrity and may be considered for more evaluations (Mahboubi et al., 2018). Based on hub and bottleneck definition, when the function of one gene impairs, it may dramatically affect the function of other linked genes. Network analysis in Figure 1 (A & B) reveals that the location of the hub and bottleneck elements are well-defined in the right down region of degree and top region of BC distribution plots, respectively. Centrality analysis implies the significant contribution of ESR1, DRD2, HTR1B, DRD4, TNFA, HTR2A, and CDH2 in the OCD network. In the next step, a backbone network of central genes revealed 7 essential genes whose pivotal role in network integrity requires further analysis. About 83% of the nodes of the main network are presented in the backbone network. However, about 70% of the edges are constructed. As it is tabulated in Table 3, cross-validation confirms the robustness of the network since the frequency of hub-bottleneck genes was not affected by the changes in the initial genes. Therefore, the backbone network shows excellent accuracy for four deletions, and the pivotal role of hub-bottleneck genes is supported. According to Table 3 and Figure 2, the accuracy values vary from 95% to 98%. Besides, detailed characteristic of each vital elements as collected in Table 4, indicates the importance of these genes in OCD subtypes. In this light, ESR1 as the top hub-bottleneck gene in the network may play a major role in OCD. On the other hand, studies showed that ESR1 is associated with other mental disorders as well. ESR1 is seen in the several regions of the brain that are connected to cognitive function and emotional behavior. Steroid hormones regulate compulsive behavior, mood, and cognition (Sundermann, Maki, & Bishop, 2010). Estrogen hormone has a critical regulatory activity on serototonergic, glutamate, and dopaminergic pathways (Alonso et al., 2011). Some kinds of variants in ESR1 can cause changes in estrogen responsivity in the brain (Rettberg, Yao, & Brinton, 2014), resulting in the manifestation of many kinds of neuropsychiatric conditions (Sundermann et al., 2010).
On the other hand, the critical associations of ESR1 polymorphisms in women’s neuropsychiatric disorders are suggested by many studies (Sundermann et al., 2010; Millet et al., 2003). Besides, gender plays a vital role in OCD phenotype (Mathis et al., 2011). Women are more susceptible to certain types of OCD than men. For instance, cleaning obsessions and contamination compulsions are more evident in females that may be connected to ESR1 malfunction (Nissen et al., 2016). It is also suggested by many clinical assessments that there is a vital connection between OCD onset and severity and reproductive cycle activity, in which the level of hormones varies greatly (Vulink, Denys, Bus, & Westenberg, 2006; Labad, Menchón, & Alonso, 2005; Forray, Focseneanu, Pittman, McDougle, & Epperson, 2010; Guglielmi et al., 2014). It seems that ESR1, as part of reproductive cycle events, may play a key role in the pathogenesis of OCD.
Moreover, its central role in the OCD network reflects the possible crucial role of female reproductive cycle activity in OCD onset and development. TNF-α, the second essential gene in the network, has a noticeable function in immunologic reactions that suggests the vital contribution of immunologic reactions in OCD risk. It is also reported in previous investigations that TNF-α has an essential role in some subtypes of OCD (Hounie et al., 2008). DRD2 and DRD4 also implied key roles in the OCD network. It is also confirmed by previous studies that the dopamine system can be an appropriate target for OCD treatment. HTR1B and HTR2A are serotonin receptors, and their relation to OCD is gender-specific. HTR1B has a remarkable contribution in male patients (Mas et al., 2014), while HTR2A is mostly involved in females (Enoch et al., 2001). Another central gene, CDH2, has a primary role in early brain morphogenesis, long-term potentiation, synaptic plasticity, and synaptogenesis. This gene, as a producer of cell-cell adhesion protein, belongs to the cadherin gene superfamily. In OCD, dysregulation of the glutamate receptor availability may attribute to CDH2 polymorphisms (McGregor et al., 2016).
According to the ClueGO findings (Figure 3 and Table 5), dopamine transport, learning, memory, and monoamine transport processes may play an essential role in OCD etiology. These BPs may be influenced by the different contributing polymorphisms. Most genes in these processes are among the ranked genes. The highest-ranked process in this analysis is dopamine transport. On the other hand, most of the prescribed medicines for OCD belong to the Selective Serotonin Reuptake Inhibitors (SSRIs) family. However, not all patients’ responses to this drug family and other treatment options are essential to be evaluated. One of the potential therapeutic targets is the dopamine system; dopaminergic antagonists are designed and showed promising treatment-resistant cases (Koo et al., 2010). Here, the contribution of four central genes in the dopamine transport process suggests the important role of this process in OCD pathogenesis. Another nominated critical process in OCD pathogenesis is learning, suggesting that OCD patients may have impairments in learning. It has been reported that patients with OCD are slower in learning (Gross-Isseroff et al., 1996). Memory is another involved process in OCD seed genes analysis. This fact can be justified by previous clinical findings of memory impairments in OCD (Penadés, Catalán, Andrés, Salamero, & Gastó, 2005). In other words, our analysis provides more supports for memory process dysregulation in OCD. Overall, the investigation introduced some feasible candidates showing substantial participation in our network integrity and functions. Variation in OCD genes may lead to the formation of different combinations of genes’ functions. As a result, each of these combinations can correspond to a specific OCD subtype model. Activation of a particular combination is attributed to a specific internal or external stimulation. Hence, the onset of a specific subgroup for each individual depends on the origin of the trigger and the influenced pathway.
Our study results suggest topological information of interactome profile of OCD reported genes from genetic studies; however, it is in its initial phase. Our PPI network analysis can serve as a paradigm for later experiments such as genomic, proteomic, and metabolomic investigations. Thus, by enriching interaction sets, a more in-depth understanding of OCD etiology may be reachable. Finally, screening 31 genes associated with OCD leads to introducing a panel, including 7 crucial genes. This panel may potentially be used for diagnostic and therapeutic proposes of OCD after extensive validation tests.
Ethical Considerations
Compliance with ethical guidelines
All ethical principles are considered in this article.
Funding
This study was supported by the Proteomics Research center, Tehran.
Authors' contributions
All authors contributed equally to work.
Conflict of interest
The authors declared no conflict of interest.
five times by selecting one hub-bottleneck gene and randomly selecting from other top genes (Table 3, Figure 2).
A backbone network was built from Mentha, Reactome, and Reactome-Fls sources for 7 hub-bottleneck elements. This network has 1609 nodes, and 2279 edges denote that 83% of the nodes relative to the main network are presented in the backbone network.
Different subtypes have been reported for OCD that may appear due to the interaction of different genes and environmental factors. Central genes in this study are reported with different subtypes (Table 4).
Application of ClueGO plugin for functional enrichment analysis of OCD seed genes based on BP terms is shown in Figure 3 and Table 5.
Groups are labeled based on the leading term (the most significant). The most significant term is defined as the term with the highest number and percentage of initial genes. Kappa Score ≥0.5. Figure 3 showed that dopamine transport, learning, memory, and monoamine transport are the main BPs.
4. Discussion
Molecular studies of Obsessive-Compulsive Disorder (OCD) have just begun (Zamanian-Azodi et al., 2015). Most of the recent investigations are focused on the genetic basis of OCD. Multiple genetic variants with small effect size are linked to OCD risk (Gruenblatt, Hauser, & Walitza, 2014). While individual examining of genes is informative, interactome evaluation is essential as well. Genes are in a complex interaction structure that any genetic alterations may lead to functional phenotype changes of the whole system (Chavez et al., 2013). PPI network analysis can identify specific interactome changes in a disease state. In this regard, examining the OCD interactome pattern can help gain further insight into the underlying mechanisms through topological analysis (Mahboubi et al., 2018). In this study, a total number of 31 human candidate genes from genetic association studies are reviewed. Among them, 8 genes were previously reported significant for OCD by a meta-analysis (Taylor, 2013).
On the other hand, none of the single candidate genes has been proved to be significantly associated with OCD by genome-wide association studies (Gruenblatt et al., 2014). Moreover, mRNA expression profiling of postmortem brain of OCD patients showed no significant associations of these genes and mRNA expression profile (Jaffe et al., 2014). Therefore, a network from reported genes may be inaccurate since they are incomplete and may show some bias towards neurotransmitter systems genes (Gruenblatt et al., 2014). To somehow overcome this limitation, it is tried to select genes from genetic-based studies that were with at least one significant association report with OCD.
There are about 1.5 edges per node; however, this value’s variation is considerable for different nodes. For example, ESR1, the top node, is characterized by 899 links. This gene is discriminated by about 650 links from the second top score node. In a PPI network, central genes (hub-bottlenecks) are essential for network integrity and may be considered for more evaluations (Mahboubi et al., 2018). Based on hub and bottleneck definition, when the function of one gene impairs, it may dramatically affect the function of other linked genes. Network analysis in Figure 1 (A & B) reveals that the location of the hub and bottleneck elements are well-defined in the right down region of degree and top region of BC distribution plots, respectively. Centrality analysis implies the significant contribution of ESR1, DRD2, HTR1B, DRD4, TNFA, HTR2A, and CDH2 in the OCD network. In the next step, a backbone network of central genes revealed 7 essential genes whose pivotal role in network integrity requires further analysis. About 83% of the nodes of the main network are presented in the backbone network. However, about 70% of the edges are constructed. As it is tabulated in Table 3, cross-validation confirms the robustness of the network since the frequency of hub-bottleneck genes was not affected by the changes in the initial genes. Therefore, the backbone network shows excellent accuracy for four deletions, and the pivotal role of hub-bottleneck genes is supported. According to Table 3 and Figure 2, the accuracy values vary from 95% to 98%. Besides, detailed characteristic of each vital elements as collected in Table 4, indicates the importance of these genes in OCD subtypes. In this light, ESR1 as the top hub-bottleneck gene in the network may play a major role in OCD. On the other hand, studies showed that ESR1 is associated with other mental disorders as well. ESR1 is seen in the several regions of the brain that are connected to cognitive function and emotional behavior. Steroid hormones regulate compulsive behavior, mood, and cognition (Sundermann, Maki, & Bishop, 2010). Estrogen hormone has a critical regulatory activity on serototonergic, glutamate, and dopaminergic pathways (Alonso et al., 2011). Some kinds of variants in ESR1 can cause changes in estrogen responsivity in the brain (Rettberg, Yao, & Brinton, 2014), resulting in the manifestation of many kinds of neuropsychiatric conditions (Sundermann et al., 2010).
On the other hand, the critical associations of ESR1 polymorphisms in women’s neuropsychiatric disorders are suggested by many studies (Sundermann et al., 2010; Millet et al., 2003). Besides, gender plays a vital role in OCD phenotype (Mathis et al., 2011). Women are more susceptible to certain types of OCD than men. For instance, cleaning obsessions and contamination compulsions are more evident in females that may be connected to ESR1 malfunction (Nissen et al., 2016). It is also suggested by many clinical assessments that there is a vital connection between OCD onset and severity and reproductive cycle activity, in which the level of hormones varies greatly (Vulink, Denys, Bus, & Westenberg, 2006; Labad, Menchón, & Alonso, 2005; Forray, Focseneanu, Pittman, McDougle, & Epperson, 2010; Guglielmi et al., 2014). It seems that ESR1, as part of reproductive cycle events, may play a key role in the pathogenesis of OCD.
Moreover, its central role in the OCD network reflects the possible crucial role of female reproductive cycle activity in OCD onset and development. TNF-α, the second essential gene in the network, has a noticeable function in immunologic reactions that suggests the vital contribution of immunologic reactions in OCD risk. It is also reported in previous investigations that TNF-α has an essential role in some subtypes of OCD (Hounie et al., 2008). DRD2 and DRD4 also implied key roles in the OCD network. It is also confirmed by previous studies that the dopamine system can be an appropriate target for OCD treatment. HTR1B and HTR2A are serotonin receptors, and their relation to OCD is gender-specific. HTR1B has a remarkable contribution in male patients (Mas et al., 2014), while HTR2A is mostly involved in females (Enoch et al., 2001). Another central gene, CDH2, has a primary role in early brain morphogenesis, long-term potentiation, synaptic plasticity, and synaptogenesis. This gene, as a producer of cell-cell adhesion protein, belongs to the cadherin gene superfamily. In OCD, dysregulation of the glutamate receptor availability may attribute to CDH2 polymorphisms (McGregor et al., 2016).
According to the ClueGO findings (Figure 3 and Table 5), dopamine transport, learning, memory, and monoamine transport processes may play an essential role in OCD etiology. These BPs may be influenced by the different contributing polymorphisms. Most genes in these processes are among the ranked genes. The highest-ranked process in this analysis is dopamine transport. On the other hand, most of the prescribed medicines for OCD belong to the Selective Serotonin Reuptake Inhibitors (SSRIs) family. However, not all patients’ responses to this drug family and other treatment options are essential to be evaluated. One of the potential therapeutic targets is the dopamine system; dopaminergic antagonists are designed and showed promising treatment-resistant cases (Koo et al., 2010). Here, the contribution of four central genes in the dopamine transport process suggests the important role of this process in OCD pathogenesis. Another nominated critical process in OCD pathogenesis is learning, suggesting that OCD patients may have impairments in learning. It has been reported that patients with OCD are slower in learning (Gross-Isseroff et al., 1996). Memory is another involved process in OCD seed genes analysis. This fact can be justified by previous clinical findings of memory impairments in OCD (Penadés, Catalán, Andrés, Salamero, & Gastó, 2005). In other words, our analysis provides more supports for memory process dysregulation in OCD. Overall, the investigation introduced some feasible candidates showing substantial participation in our network integrity and functions. Variation in OCD genes may lead to the formation of different combinations of genes’ functions. As a result, each of these combinations can correspond to a specific OCD subtype model. Activation of a particular combination is attributed to a specific internal or external stimulation. Hence, the onset of a specific subgroup for each individual depends on the origin of the trigger and the influenced pathway.
Our study results suggest topological information of interactome profile of OCD reported genes from genetic studies; however, it is in its initial phase. Our PPI network analysis can serve as a paradigm for later experiments such as genomic, proteomic, and metabolomic investigations. Thus, by enriching interaction sets, a more in-depth understanding of OCD etiology may be reachable. Finally, screening 31 genes associated with OCD leads to introducing a panel, including 7 crucial genes. This panel may potentially be used for diagnostic and therapeutic proposes of OCD after extensive validation tests.
Ethical Considerations
Compliance with ethical guidelines
All ethical principles are considered in this article.
Funding
This study was supported by the Proteomics Research center, Tehran.
Authors' contributions
All authors contributed equally to work.
Conflict of interest
The authors declared no conflict of interest.
References
Alonso, P., Gratacos, M., Menchon, J. M., Segalas, C., Gonzalez, J. R., & Labad, J., et al. (2008). Genetic susceptibility to obsessive-compulsive hoarding: The contribution of neurotrophic tyrosine kinase receptor type 3 gene 1. Genes, Brain and Behavior, 7(7), 778-85. [DOI:10.1111/j.1601-183X.2008.00418.x] [PMID]
Alonso, P., Gratacos, M., Segalas, C., Escaramis, G., Real, E., & Bayes, M., et al. (2011). Variants in estrogen receptor alpha gene are associated with phenotypical expression of obsessive-compulsive disorder. Psychoneuroendocrinology, 36(4), 473-83. [DOI:10.1016/j.psyneuen.2010.07.022] [PMID]
Aranda, B., Blankenburg, H., Kerrien, S., Brinkman, F. S., Ceol, A., & Chautard, E., et al. (2011). PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nature Methods, 8(7), 528-9. [DOI:10.1038/nmeth.1637] [PMID]
Arnold, P. D., Rosenberg, D. R., Mundo, E., Tharmalingam, S., Kennedy, J. L., & Richter, M. A. (2004). Association of a glutamate (NMDA) subunit receptor gene (GRIN2B) with obsessive-compulsive disorder: A preliminary study. Psychopharmacology, 174(4), 530-8. [DOI:10.1007/s00213-004-1847-1] [PMID]
Assenov, Y., Ramírez, F., Schelhorn, S. E., Lengauer, T., & Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics, 24(2), 282-4. [DOI:10.1093/bioinformatics/btm554] [PMID]
Bindea, G., Galon, J., & Mlecnik, B. (2013). CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics, 29(5), 661-3. [DOI:10.1093/bioinformatics/btt019] [PMID]
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., & Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091-3. [DOI:10.1093/bioinformatics/btp101] [PMID]
Boardman, L., van der Merwe, L., Lochner, C., Kinnear, C. J., Seedat, S., & Stein, D. J., et al. (2011). Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Comprehensive Psychiatry, 52(2), 181-7. [DOI:10.1016/j.comppsych.2010.05.007] [PMID]
Chavez, J. D., Weisbrod, C. R., Zheng, C., Eng, J. K., & Bruce, J. E. (2013). Protein interactions, post-translational modifications and topologies in human cells. Molecular & Cellular Proteomics, 12(5), 1451-67. [DOI:10.1074/mcp.M112.024497] [PMID]
Enoch, M. A., Greenberg, B. D., Murphy, D. L., & Goldman, D. (2001). Sexually dimorphic relationship of a 5-HT2A promoter polymorphism with obsessive-compulsive disorder. Biological Psychiatry, 49(4), 385-8. [DOI:10.1016/S0006-3223(00)01040-4]
Forray, A., Focseneanu, M., Pittman, B., McDougle, C. J., & Epperson, C. N. (2010). Onset and exacerbation of obsessive-compulsive disorder in pregnancy and the postpartum period. The Journal of Clinical Psychiatry, 71(8), 1061-68. [DOI:10.4088/JCP.09m05381blu] [PMID]
Gross-Isseroff, R., Sasson, Y., Voet, H., Hendler, T., Luca-Haimovici, K., & Kandel-Sussman, H., et al. (1996). Alternation learning in obsessive-compulsive disorder. Biological Psychiatry, 39(8), 733-8. [DOI:10.1016/0006-3223(95)00179-4]
Gruenblatt, E., Hauser, T. U., & Walitza, S. (2014). Imaging genetics in obsessive-compulsive disorder: linking genetic variations to alterations in neuroimaging. Progress in Neurobiology, 121, 114-24. [DOI:10.1016/j.pneurobio.2014.07.003] [PMID]
Guglielmi, V., Vulink, N. C., Denys, D., Wang, Y., Samuels, J. F., & Nestadt, G. (2014). Obsessive–compulsive disorder and female reproductive cycle events: Results from the OCD and reproduction collaborative study. Depression and Anxiety, 31(12), 979-87. [DOI:10.1002/da.22234] [PMID]
Hounie, A. G., Cappi, C., Cordeiro, Q., Sampaio, A. S., Moraes, I., & do Rosario, M. C., et al. (2008). TNF-alpha polymorphisms are associated with obsessive-compulsive disorder. Neuroscience Letters, 442(2), 86-90. [DOI:10.1016/j.neulet.2008.07.022] [PMID]
Ickowicz, A., Feng, Y., Wigg, K., Quist, J., Pathare, T., & Roberts, W., et al. (2007). The serotonin receptor HTR1B: Gene polymorphisms in attention deficit hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 144(1), 121-5. [DOI:10.1002/ajmg.b.30398] [PMID]
Jaffe, A. E., Deep-Soboslay, A., Tao, R., Hauptman, D. T., Kaye, W. H., & Arango, V., et al. (2014). Genetic neuropathology of obsessive psychiatric syndromes. Translational Psychiatry, 4(9), e432. [DOI:10.1038/tp.2014.68] [PMID]
Koo, M. S., Kim, E. J., Roh, D., & Kim, C. H. (2010). Role of dopamine in the pathophysiology and treatment of obsessive–compulsive disorder. Expert Review of Neurotherapeutics, 10(2), 275-90. [DOI:10.1586/ern.09.148] [PMID]
Labad, J., Menchón, J. M., & Alonso, P. (2005). Female reproductive cycle and obsessive-compulsive disorder. The Journal of Clinical Psychiatry, 66, 428-35. [DOI:10.4088/JCP.v66n0404] [PMID]
Lazar, A., Walitza, S., Jetter, A., Gerlach, M., Warnke, A., & Herpertz-Dahlmann, B., et al. (2008). Novel mutations of the extraneuronal monoamine transporter gene in children and adolescents with obsessive–compulsive disorder. International Journal of Neuropsychopharmacology, 11(1), 35-48. [DOI:10.1017/S1461145707007742] [PMID]
Lennertz, L., Wagner, M., Grabe, H. J., Franke, P. E., Guttenthaler, V., & Rampacher, F., et al. (2014). 5-HT3 receptor influences the washing phenotype and visual organization in obsessive-compulsive disorder supporting 5-HT3 receptor antagonists as novel treatment option. European Neuropsychopharmacology, 24(1), 86-94. [DOI:10.1016/j.euroneuro.2013.07.003] [PMID]
Li, J., Cui, J., Wang, X., Ma, J., Niu, H., & Ma, X., et al. (2015). An association study between DLGAP1 rs11081062 and EFNA5 rs26728 polymorphisms with obsessive–compulsive disorder in a Chinese Han population. Neuropsychiatric Disease and Treatment, 11, 897. [DOI:10.2147/NDT.S75009] [PMID]
Lochner, C., McGregor, N., Hemmings, S., Harvey, B. H., Breet, E., & Swanevelder, S., et al. (2016). Symmetry symptoms in obsessive-compulsive disorder: Clinical and genetic correlates. Brazilian Journal of Psychiatry, 38(1), 17-23. [DOI:10.1590/1516-4446-2014-1619] [PMID]
Lotia, S., Montojo, J., Dong, Y., Bader, G. D., & Pico, A. R. (2013). Cytoscape app store. Bioinformatics, 29(10), 1350-1. [DOI:10.1093/bioinformatics/btt138] [PMID]
Luo, X., Huang, L., Jia, P., Li, M., Su, B., & Zhao, Z., et al. (2014). Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophrenia Bulletin, 40(1), 39-49. [DOI:10.1093/schbul/sbt066] [PMID]
Mahboubi, M., Azodi, M. Z., Tavirani, M. R., Mansouri, V., Ahmadi, N. A., & Hamdieh, M., et al. (2018). Protein-Protein Interaction Analysis of Common Top Genes in Obsessive-Compulsive Disorder (OCD) and Schizophrenia: Towards New Drug Approach Obsessive-Compulsive disorder (OCD) and Schizophrenia Comorbidity Gene Analysis. Iranian Journal of Pharmaceutical Research: IJPR, 17(Suppl2), 173-86. [PMCID] [PMID]
Mas, S., Pagerols, M., Gassó, P., Ortiz, A., Rodriguez, N., & Morer, A., et al. (2014). Role of GAD2 and HTR1B genes in early-onset obsessive-compulsive disorder: Results from transmission disequilibrium study. Genes, Brain and Behavior, 13(4), 409-17. [DOI:10.1111/gbb.12128] [PMID]
Mathis, M. A. D., Alvarenga, P. D., Funaro, G., Torresan, R. C., Moraes, I., & Torres, A. R., et al. (2011). Gender differences in obsessive-compulsive disorder: A literature review. Brazilian Journal of Psychiatry, 33(4), 390-9.[DOI:10.1590/S1516-44462011000400014] [PMID]
McGregor, N. W., Lochner, C., Stein, D. J., & Hemmings, S. M. J. (2016). Polymorphisms within the neuronal cadherin (CDH2) gene are associated with obsessive-compulsive disorder (OCD) in a South African cohort. Metabolic Brain Disease, 31(1), 191-6. [DOI:10.1007/s11011-015-9693-x] [PMID]
Millet, B., Chabane, N., Delorme, R., Leboyer, M., Leroy, S., & Poirier, M. F., et al. (2003). Association between the dopamine receptor D4 (DRD4) gene and obsessive-compulsive disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 116(1), 55-9. [DOI:10.1002/ajmg.b.10034] [PMID]
Walitza, S., Renner, T. J., Dempfle, A., Konrad, K., Wewetzer, C., & Halbach, A., et al. (2005). Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Molecular Psychiatry, 10(12), 1126-32. [DOI:10.1017/S1461145705005997] [PMID]
Mundo, E., Richter, M. A., Sam, F., Macciardi, F., & Kennedy, J. L. (2000). Is the 5-HT1Dβ receptor gene implicated in the pathogenesis of obsessive-compulsive disorder? American Journal of Psychiatry, 157(7), 1160-1. [DOI:10.1176/appi.ajp.157.7.1160] [PMID]
Nair, J., Ghatge, M., Kakkar, V. V., & Shanker, J. (2014). Network analysis of inflammatory genes and their transcriptional regulators in coronary artery disease. PloS One, 9(4), e94328. [DOI:10.1371/journal.pone.0094328] [PMID]
Nicolini, H., Cruz, C., Camarena, B., Orozco, B., Kennedy, J. L., & King, N., et al. (1996). DRD2, DRD3 and 5HT2A receptor genes polymorphisms in obsessive-compulsive disorder. Mol Psychiatry, 1(6), 461-5. https://www.researchgate.net/profile/Beatriz-
Camarena/publication/14064809_DRD2_DRD3_and_5HT2A_receptor_genes_polymorphisms_in_ObsessiveCompulsive_Disorder/links/547f52dd0cf25b80dd6e5206/DRD2-DRD3-and-5HT2A-receptor-genes-polymorphisms-in-Obsessive-Compulsive-Disorder.pdf
Nissen, J. B., Hansen, C. S., Starnawska, A., Mattheisen, M., Børglum, A. D., & Buttenschøn, H. N., et al. (2016). DNA methylation at the neonatal state and at the time of diagnosis: preliminary support for an association with the estrogen receptor 1, gamma-aminobutyric acid B receptor 1, and myelin oligodendrocyte glycoprotein in female adolescent patients with OCD. Frontiers in Psychiatry, 7, 35. [DOI:10.3389/fpsyt.2016.00035]
Ozomaro, U., Cai, G., Kajiwara, Y., Yoon, S., Makarov, V., & Delorme, R., et al. (2013). Characterization of SLITRK1 variation in obsessive-compulsive disorder. PloS One, 8(8), e70376. [DOI:10.1371/journal.pone.0070376] [PMID]
Penadés, R., Catalán, R., Andrés, S., Salamero, M., & Gastó, C. (2005). Executive function and nonverbal memory in obsessive-compulsive disorder. Psychiatry Research, 133(1), 81-90. [DOI:10.1016/j.psychres.2004.09.005] [PMID]
Rettberg, J. R., Yao, J., & Brinton, R. D. (2014). Estrogen: a master regulator of bioenergetic systems in the brain and body. Frontiers in Neuroendocrinology, 35(1), 8-30. [DOI:10.1016/j.yfrne.2013.08.001] [PMID]
Rezaei-Tavirani, M., Tavirani, M. R., & Azodi, M. Z. (2019). The bioinformatics aspects of gene screening of HT-29, human colon cell line treated with caffeic acid. Gastroenterology and Hepatology From Bed to Bench, 12(3), 246. [DOI:10.5812/ijcm.92465]
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., & Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nature Methods, 9(11), 1069. [DOI:10.1038/nmeth.2212] [PMID]
Scardoni, G., Petterlini, M., & Laudanna, C. (2009). Analyzing biological network parameters with CentiScaPe. Bioinformatics, 25(21), 2857-9. [DOI:10.1093/bioinformatics/btp517] [PMID]
Stewart, S. E., Mayerfeld, C., Arnold, P. D., Crane, J. R., O'Dushlaine, C., & Fagerness, J. A., et al. (2013). Meta-analysis of association between obsessive-compulsive disorder and the 3′ region of neuronal glutamate transporter gene SLC1A1. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 162(4), 367-9. [DOI:10.1002/ajmg.b.32137] [PMID]
Sundermann, E. E., Maki, P. M., & Bishop, J. R. (2010). A review of Estrogen Receptor Α Gene (ESR1) polymorphisms, mood, and cognition. Menopause (New York, NY), 17(4), 874. [DOI:10.1097/gme.0b013e3181df4a19] [PMID]
Tang, R., Noh, H. J., Wang, D., Sigurdsson, S., Swofford, R., & Perloski, M., et al. (2014). Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biology, 15(3), 1-15. [DOI:10.1186/gb-2014-15-3-r25] [PMID]
Taylor, S. (2013). Molecular genetics of obsessive–compulsive disorder: A comprehensive meta-analysis of genetic association studies. Molecular Psychiatry, 18(7), 799-805. [DOI:10.1038/mp.2012.76] [PMID]
Taylor, S., Asmundson, G. J., & Jang, K. L. (2016). Etiology of obsessions and compulsions: General and specific genetic and environmental factors. Psychiatry Research, 237, 17-21. [DOI:10.1016/j.psychres.2016.01.071] [PMID]
Viswanath, B., Purushottam, M., Kandavel, T., Reddy, Y. J., & Jain, S. (2013). DRD4 gene and obsessive compulsive disorder: Do symptom dimensions have specific genetic correlates? Progress in Neuro-Psychopharmacology and Biological Psychiatry, 41, 18-23. [DOI:10.1016/j.pnpbp.2012.10.023] [PMID]
Vulink, N. C., Denys, D., Bus, L., & Westenberg, H. G. (2006). Female hormones affect symptom severity in obsessive–compulsive disorder. International Clinical Psychopharmacology, 21(3), 171-5. [DOI:10.1097/01.yic.0000199454.62423.99] [PMID]
Zai, G., Bezchlibnyk, Y. B., Richter, M. A., Arnold, P., Burroughs, E., Barr, C. L., & Kennedy, J. L. (2004). Myelin Oligodendrocyte Glycoprotein (MOG) gene is associated with obsessive-compulsive disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 129(1), 64-8. [DOI:10.1002/ajmg.b.30077] [PMID]
Zai, G., Zai, C. C., Arnold, P. D., Freeman, N., Burroughs, E., & Kennedy, J. L., et al. (2015). Meta-analysis and association of Brain-Derived Neurotrophic Factor (BDNF) gene with obsessive-compulsive disorder. Psychiatric Genetics, 25(2), 95-6. [DOI:10.1097/YPG.0000000000000077] [PMID]
Zamanian-Azodi, M., Rezaei-Tavirani, M., Kermani-Ranjbar, T., Arefi Oskouie, A., Rezaei-Tavirani, M., & RahmatiRad, S., et al. (2015). [Pathophysiology, genetics, types, and treatments in obsessive compulsive disorder (Persian)]. Koomesh, 16(4), 475-87. http://eprints.semums.ac.ir/230/1/Koomesh%2Dv16n4p475%2Den.pdf
Zamanian-Azodi, M., Rezaei-Tavirani, M., Mahboubi, M., Hamidpour, M., Tavirani, M. R., & Hamdieh, M., et al. (2018). Serum proteomic study of women with obsessive-compulsive disorder, washing subtype. Basic and Clinical Neuroscience, 9(5), 337. [DOI:10.32598/bcn.9.5.337] [PMID]
Alonso, P., Gratacos, M., Menchon, J. M., Segalas, C., Gonzalez, J. R., & Labad, J., et al. (2008). Genetic susceptibility to obsessive-compulsive hoarding: The contribution of neurotrophic tyrosine kinase receptor type 3 gene 1. Genes, Brain and Behavior, 7(7), 778-85. [DOI:10.1111/j.1601-183X.2008.00418.x] [PMID]
Alonso, P., Gratacos, M., Segalas, C., Escaramis, G., Real, E., & Bayes, M., et al. (2011). Variants in estrogen receptor alpha gene are associated with phenotypical expression of obsessive-compulsive disorder. Psychoneuroendocrinology, 36(4), 473-83. [DOI:10.1016/j.psyneuen.2010.07.022] [PMID]
Aranda, B., Blankenburg, H., Kerrien, S., Brinkman, F. S., Ceol, A., & Chautard, E., et al. (2011). PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nature Methods, 8(7), 528-9. [DOI:10.1038/nmeth.1637] [PMID]
Arnold, P. D., Rosenberg, D. R., Mundo, E., Tharmalingam, S., Kennedy, J. L., & Richter, M. A. (2004). Association of a glutamate (NMDA) subunit receptor gene (GRIN2B) with obsessive-compulsive disorder: A preliminary study. Psychopharmacology, 174(4), 530-8. [DOI:10.1007/s00213-004-1847-1] [PMID]
Assenov, Y., Ramírez, F., Schelhorn, S. E., Lengauer, T., & Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics, 24(2), 282-4. [DOI:10.1093/bioinformatics/btm554] [PMID]
Bindea, G., Galon, J., & Mlecnik, B. (2013). CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics, 29(5), 661-3. [DOI:10.1093/bioinformatics/btt019] [PMID]
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., & Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091-3. [DOI:10.1093/bioinformatics/btp101] [PMID]
Boardman, L., van der Merwe, L., Lochner, C., Kinnear, C. J., Seedat, S., & Stein, D. J., et al. (2011). Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Comprehensive Psychiatry, 52(2), 181-7. [DOI:10.1016/j.comppsych.2010.05.007] [PMID]
Chavez, J. D., Weisbrod, C. R., Zheng, C., Eng, J. K., & Bruce, J. E. (2013). Protein interactions, post-translational modifications and topologies in human cells. Molecular & Cellular Proteomics, 12(5), 1451-67. [DOI:10.1074/mcp.M112.024497] [PMID]
Enoch, M. A., Greenberg, B. D., Murphy, D. L., & Goldman, D. (2001). Sexually dimorphic relationship of a 5-HT2A promoter polymorphism with obsessive-compulsive disorder. Biological Psychiatry, 49(4), 385-8. [DOI:10.1016/S0006-3223(00)01040-4]
Forray, A., Focseneanu, M., Pittman, B., McDougle, C. J., & Epperson, C. N. (2010). Onset and exacerbation of obsessive-compulsive disorder in pregnancy and the postpartum period. The Journal of Clinical Psychiatry, 71(8), 1061-68. [DOI:10.4088/JCP.09m05381blu] [PMID]
Gross-Isseroff, R., Sasson, Y., Voet, H., Hendler, T., Luca-Haimovici, K., & Kandel-Sussman, H., et al. (1996). Alternation learning in obsessive-compulsive disorder. Biological Psychiatry, 39(8), 733-8. [DOI:10.1016/0006-3223(95)00179-4]
Gruenblatt, E., Hauser, T. U., & Walitza, S. (2014). Imaging genetics in obsessive-compulsive disorder: linking genetic variations to alterations in neuroimaging. Progress in Neurobiology, 121, 114-24. [DOI:10.1016/j.pneurobio.2014.07.003] [PMID]
Guglielmi, V., Vulink, N. C., Denys, D., Wang, Y., Samuels, J. F., & Nestadt, G. (2014). Obsessive–compulsive disorder and female reproductive cycle events: Results from the OCD and reproduction collaborative study. Depression and Anxiety, 31(12), 979-87. [DOI:10.1002/da.22234] [PMID]
Hounie, A. G., Cappi, C., Cordeiro, Q., Sampaio, A. S., Moraes, I., & do Rosario, M. C., et al. (2008). TNF-alpha polymorphisms are associated with obsessive-compulsive disorder. Neuroscience Letters, 442(2), 86-90. [DOI:10.1016/j.neulet.2008.07.022] [PMID]
Ickowicz, A., Feng, Y., Wigg, K., Quist, J., Pathare, T., & Roberts, W., et al. (2007). The serotonin receptor HTR1B: Gene polymorphisms in attention deficit hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 144(1), 121-5. [DOI:10.1002/ajmg.b.30398] [PMID]
Jaffe, A. E., Deep-Soboslay, A., Tao, R., Hauptman, D. T., Kaye, W. H., & Arango, V., et al. (2014). Genetic neuropathology of obsessive psychiatric syndromes. Translational Psychiatry, 4(9), e432. [DOI:10.1038/tp.2014.68] [PMID]
Koo, M. S., Kim, E. J., Roh, D., & Kim, C. H. (2010). Role of dopamine in the pathophysiology and treatment of obsessive–compulsive disorder. Expert Review of Neurotherapeutics, 10(2), 275-90. [DOI:10.1586/ern.09.148] [PMID]
Labad, J., Menchón, J. M., & Alonso, P. (2005). Female reproductive cycle and obsessive-compulsive disorder. The Journal of Clinical Psychiatry, 66, 428-35. [DOI:10.4088/JCP.v66n0404] [PMID]
Lazar, A., Walitza, S., Jetter, A., Gerlach, M., Warnke, A., & Herpertz-Dahlmann, B., et al. (2008). Novel mutations of the extraneuronal monoamine transporter gene in children and adolescents with obsessive–compulsive disorder. International Journal of Neuropsychopharmacology, 11(1), 35-48. [DOI:10.1017/S1461145707007742] [PMID]
Lennertz, L., Wagner, M., Grabe, H. J., Franke, P. E., Guttenthaler, V., & Rampacher, F., et al. (2014). 5-HT3 receptor influences the washing phenotype and visual organization in obsessive-compulsive disorder supporting 5-HT3 receptor antagonists as novel treatment option. European Neuropsychopharmacology, 24(1), 86-94. [DOI:10.1016/j.euroneuro.2013.07.003] [PMID]
Li, J., Cui, J., Wang, X., Ma, J., Niu, H., & Ma, X., et al. (2015). An association study between DLGAP1 rs11081062 and EFNA5 rs26728 polymorphisms with obsessive–compulsive disorder in a Chinese Han population. Neuropsychiatric Disease and Treatment, 11, 897. [DOI:10.2147/NDT.S75009] [PMID]
Lochner, C., McGregor, N., Hemmings, S., Harvey, B. H., Breet, E., & Swanevelder, S., et al. (2016). Symmetry symptoms in obsessive-compulsive disorder: Clinical and genetic correlates. Brazilian Journal of Psychiatry, 38(1), 17-23. [DOI:10.1590/1516-4446-2014-1619] [PMID]
Lotia, S., Montojo, J., Dong, Y., Bader, G. D., & Pico, A. R. (2013). Cytoscape app store. Bioinformatics, 29(10), 1350-1. [DOI:10.1093/bioinformatics/btt138] [PMID]
Luo, X., Huang, L., Jia, P., Li, M., Su, B., & Zhao, Z., et al. (2014). Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophrenia Bulletin, 40(1), 39-49. [DOI:10.1093/schbul/sbt066] [PMID]
Mahboubi, M., Azodi, M. Z., Tavirani, M. R., Mansouri, V., Ahmadi, N. A., & Hamdieh, M., et al. (2018). Protein-Protein Interaction Analysis of Common Top Genes in Obsessive-Compulsive Disorder (OCD) and Schizophrenia: Towards New Drug Approach Obsessive-Compulsive disorder (OCD) and Schizophrenia Comorbidity Gene Analysis. Iranian Journal of Pharmaceutical Research: IJPR, 17(Suppl2), 173-86. [PMCID] [PMID]
Mas, S., Pagerols, M., Gassó, P., Ortiz, A., Rodriguez, N., & Morer, A., et al. (2014). Role of GAD2 and HTR1B genes in early-onset obsessive-compulsive disorder: Results from transmission disequilibrium study. Genes, Brain and Behavior, 13(4), 409-17. [DOI:10.1111/gbb.12128] [PMID]
Mathis, M. A. D., Alvarenga, P. D., Funaro, G., Torresan, R. C., Moraes, I., & Torres, A. R., et al. (2011). Gender differences in obsessive-compulsive disorder: A literature review. Brazilian Journal of Psychiatry, 33(4), 390-9.[DOI:10.1590/S1516-44462011000400014] [PMID]
McGregor, N. W., Lochner, C., Stein, D. J., & Hemmings, S. M. J. (2016). Polymorphisms within the neuronal cadherin (CDH2) gene are associated with obsessive-compulsive disorder (OCD) in a South African cohort. Metabolic Brain Disease, 31(1), 191-6. [DOI:10.1007/s11011-015-9693-x] [PMID]
Millet, B., Chabane, N., Delorme, R., Leboyer, M., Leroy, S., & Poirier, M. F., et al. (2003). Association between the dopamine receptor D4 (DRD4) gene and obsessive-compulsive disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 116(1), 55-9. [DOI:10.1002/ajmg.b.10034] [PMID]
Walitza, S., Renner, T. J., Dempfle, A., Konrad, K., Wewetzer, C., & Halbach, A., et al. (2005). Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Molecular Psychiatry, 10(12), 1126-32. [DOI:10.1017/S1461145705005997] [PMID]
Mundo, E., Richter, M. A., Sam, F., Macciardi, F., & Kennedy, J. L. (2000). Is the 5-HT1Dβ receptor gene implicated in the pathogenesis of obsessive-compulsive disorder? American Journal of Psychiatry, 157(7), 1160-1. [DOI:10.1176/appi.ajp.157.7.1160] [PMID]
Nair, J., Ghatge, M., Kakkar, V. V., & Shanker, J. (2014). Network analysis of inflammatory genes and their transcriptional regulators in coronary artery disease. PloS One, 9(4), e94328. [DOI:10.1371/journal.pone.0094328] [PMID]
Nicolini, H., Cruz, C., Camarena, B., Orozco, B., Kennedy, J. L., & King, N., et al. (1996). DRD2, DRD3 and 5HT2A receptor genes polymorphisms in obsessive-compulsive disorder. Mol Psychiatry, 1(6), 461-5. https://www.researchgate.net/profile/Beatriz-
Camarena/publication/14064809_DRD2_DRD3_and_5HT2A_receptor_genes_polymorphisms_in_ObsessiveCompulsive_Disorder/links/547f52dd0cf25b80dd6e5206/DRD2-DRD3-and-5HT2A-receptor-genes-polymorphisms-in-Obsessive-Compulsive-Disorder.pdf
Nissen, J. B., Hansen, C. S., Starnawska, A., Mattheisen, M., Børglum, A. D., & Buttenschøn, H. N., et al. (2016). DNA methylation at the neonatal state and at the time of diagnosis: preliminary support for an association with the estrogen receptor 1, gamma-aminobutyric acid B receptor 1, and myelin oligodendrocyte glycoprotein in female adolescent patients with OCD. Frontiers in Psychiatry, 7, 35. [DOI:10.3389/fpsyt.2016.00035]
Ozomaro, U., Cai, G., Kajiwara, Y., Yoon, S., Makarov, V., & Delorme, R., et al. (2013). Characterization of SLITRK1 variation in obsessive-compulsive disorder. PloS One, 8(8), e70376. [DOI:10.1371/journal.pone.0070376] [PMID]
Penadés, R., Catalán, R., Andrés, S., Salamero, M., & Gastó, C. (2005). Executive function and nonverbal memory in obsessive-compulsive disorder. Psychiatry Research, 133(1), 81-90. [DOI:10.1016/j.psychres.2004.09.005] [PMID]
Rettberg, J. R., Yao, J., & Brinton, R. D. (2014). Estrogen: a master regulator of bioenergetic systems in the brain and body. Frontiers in Neuroendocrinology, 35(1), 8-30. [DOI:10.1016/j.yfrne.2013.08.001] [PMID]
Rezaei-Tavirani, M., Tavirani, M. R., & Azodi, M. Z. (2019). The bioinformatics aspects of gene screening of HT-29, human colon cell line treated with caffeic acid. Gastroenterology and Hepatology From Bed to Bench, 12(3), 246. [DOI:10.5812/ijcm.92465]
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., & Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nature Methods, 9(11), 1069. [DOI:10.1038/nmeth.2212] [PMID]
Scardoni, G., Petterlini, M., & Laudanna, C. (2009). Analyzing biological network parameters with CentiScaPe. Bioinformatics, 25(21), 2857-9. [DOI:10.1093/bioinformatics/btp517] [PMID]
Stewart, S. E., Mayerfeld, C., Arnold, P. D., Crane, J. R., O'Dushlaine, C., & Fagerness, J. A., et al. (2013). Meta-analysis of association between obsessive-compulsive disorder and the 3′ region of neuronal glutamate transporter gene SLC1A1. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 162(4), 367-9. [DOI:10.1002/ajmg.b.32137] [PMID]
Sundermann, E. E., Maki, P. M., & Bishop, J. R. (2010). A review of Estrogen Receptor Α Gene (ESR1) polymorphisms, mood, and cognition. Menopause (New York, NY), 17(4), 874. [DOI:10.1097/gme.0b013e3181df4a19] [PMID]
Tang, R., Noh, H. J., Wang, D., Sigurdsson, S., Swofford, R., & Perloski, M., et al. (2014). Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biology, 15(3), 1-15. [DOI:10.1186/gb-2014-15-3-r25] [PMID]
Taylor, S. (2013). Molecular genetics of obsessive–compulsive disorder: A comprehensive meta-analysis of genetic association studies. Molecular Psychiatry, 18(7), 799-805. [DOI:10.1038/mp.2012.76] [PMID]
Taylor, S., Asmundson, G. J., & Jang, K. L. (2016). Etiology of obsessions and compulsions: General and specific genetic and environmental factors. Psychiatry Research, 237, 17-21. [DOI:10.1016/j.psychres.2016.01.071] [PMID]
Viswanath, B., Purushottam, M., Kandavel, T., Reddy, Y. J., & Jain, S. (2013). DRD4 gene and obsessive compulsive disorder: Do symptom dimensions have specific genetic correlates? Progress in Neuro-Psychopharmacology and Biological Psychiatry, 41, 18-23. [DOI:10.1016/j.pnpbp.2012.10.023] [PMID]
Vulink, N. C., Denys, D., Bus, L., & Westenberg, H. G. (2006). Female hormones affect symptom severity in obsessive–compulsive disorder. International Clinical Psychopharmacology, 21(3), 171-5. [DOI:10.1097/01.yic.0000199454.62423.99] [PMID]
Zai, G., Bezchlibnyk, Y. B., Richter, M. A., Arnold, P., Burroughs, E., Barr, C. L., & Kennedy, J. L. (2004). Myelin Oligodendrocyte Glycoprotein (MOG) gene is associated with obsessive-compulsive disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 129(1), 64-8. [DOI:10.1002/ajmg.b.30077] [PMID]
Zai, G., Zai, C. C., Arnold, P. D., Freeman, N., Burroughs, E., & Kennedy, J. L., et al. (2015). Meta-analysis and association of Brain-Derived Neurotrophic Factor (BDNF) gene with obsessive-compulsive disorder. Psychiatric Genetics, 25(2), 95-6. [DOI:10.1097/YPG.0000000000000077] [PMID]
Zamanian-Azodi, M., Rezaei-Tavirani, M., Kermani-Ranjbar, T., Arefi Oskouie, A., Rezaei-Tavirani, M., & RahmatiRad, S., et al. (2015). [Pathophysiology, genetics, types, and treatments in obsessive compulsive disorder (Persian)]. Koomesh, 16(4), 475-87. http://eprints.semums.ac.ir/230/1/Koomesh%2Dv16n4p475%2Den.pdf
Zamanian-Azodi, M., Rezaei-Tavirani, M., Mahboubi, M., Hamidpour, M., Tavirani, M. R., & Hamdieh, M., et al. (2018). Serum proteomic study of women with obsessive-compulsive disorder, washing subtype. Basic and Clinical Neuroscience, 9(5), 337. [DOI:10.32598/bcn.9.5.337] [PMID]
Type of Study: Original |
Subject:
Clinical Neuroscience
Received: 2020/08/23 | Accepted: 2020/12/2 | Published: 2021/03/1
Received: 2020/08/23 | Accepted: 2020/12/2 | Published: 2021/03/1
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








.PNG)
.PNG)