Volume 8, Issue 4 (July & August 2017 -- 2017)
BCN 2017, 8(4): 307-316 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Zamanian-Azodi M, Rezaei-Tavirani M, Nejadi N, Arefi Oskouie A, Zayeri F, Hamdieh M, et al . Serum Proteomic Profiling of Obsessive-Compulsive Disorder, Washing Subtype: A Preliminary Study. BCN 2017; 8 (4) :307-316
URL: http://bcn.iums.ac.ir/article-1-785-en.html
URL: http://bcn.iums.ac.ir/article-1-785-en.html
Mona Zamanian-Azodi1 
 , Mostafa Rezaei-Tavirani *2
, Mostafa Rezaei-Tavirani *2 
 , Naser Nejadi3
, Naser Nejadi3 
 , Afsaneh Arefi Oskouie2
, Afsaneh Arefi Oskouie2 
 , Faird Zayeri1
, Faird Zayeri1 
 , Mostafa Hamdieh4
, Mostafa Hamdieh4 
 , Akram Safaei2
, Akram Safaei2 
 , Majid Rezaei-Tavirani5
, Majid Rezaei-Tavirani5 
 , Alireza Ahmadzadeh2
, Alireza Ahmadzadeh2 
 , Alireza Amouzandeh-Nobaveh6
, Alireza Amouzandeh-Nobaveh6 
 , Farshad Okhovatian7
, Farshad Okhovatian7 


 , Mostafa Rezaei-Tavirani *2
, Mostafa Rezaei-Tavirani *2 
 , Naser Nejadi3
, Naser Nejadi3 
 , Afsaneh Arefi Oskouie2
, Afsaneh Arefi Oskouie2 
 , Faird Zayeri1
, Faird Zayeri1 
 , Mostafa Hamdieh4
, Mostafa Hamdieh4 
 , Akram Safaei2
, Akram Safaei2 
 , Majid Rezaei-Tavirani5
, Majid Rezaei-Tavirani5 
 , Alireza Ahmadzadeh2
, Alireza Ahmadzadeh2 
 , Alireza Amouzandeh-Nobaveh6
, Alireza Amouzandeh-Nobaveh6 
 , Farshad Okhovatian7
, Farshad Okhovatian7 

1- Student Research Committee, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
2- Proteomics Research Center, School of Allied Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
3- Department of Biochemistry, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran.
4- Taleghani Hospital, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
5- Department of Surgery, School of Medicine, Iran University of Medical Sciences, Tehran, Iran.
6- Department of Microbiology, School of Medicine, Arak University of Medical Sciences, Arak, Iran.
7- Physiotherapy Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
2- Proteomics Research Center, School of Allied Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
3- Department of Biochemistry, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran.
4- Taleghani Hospital, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
5- Department of Surgery, School of Medicine, Iran University of Medical Sciences, Tehran, Iran.
6- Department of Microbiology, School of Medicine, Arak University of Medical Sciences, Arak, Iran.
7- Physiotherapy Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Keywords: Obsessive-compulsive disorder, Washing subtype, Serum, Proteomics, Protein-Protein interaction network analysis
Full-Text [PDF 1098 kb]
| Abstract (HTML)
Full-Text:
1. Introduction
Obsessive-Compulsive Disorder (OCD) is a complex neuropsychological condition with a lifetime prevalence of 1% to 2.5% in the general population (Campos, Yoshimi, Simão, Torresan, & Torres, 2015). The World Health Organization (WHO) introduced OCD as a leading cause of non-fatal illness-related disability that primarily affects people with the age range of 15 to 44 years (Zilhão et al., 2015). This anxiety disorder is characterized by intrusive thoughts and compulsive behaviors (Carmi, Dar, Zohar, & Zangen, 2015). Dysfunction in corticostriatal circuits is the main known implicated part of brain in pathology of OCD and OC-spectrum disorders (Burguiere, Monteiro, Mallet, Feng, & Graybiel, 2015).
The complex nature of this heterogeneous disorder is described by the influence of molecular and environmental interactions (Jaffe et al., 2014). That is, the combination of many factors is responsible for manifestation of this disorder with different subtypes. The contribution of many genes with their polymorphisms expresses different subtypes with overlapped symptom dimensions. This complex clinical feature of OCD is accompanied by other related disorders (comorbidity), including Tourette syndrome, chronic hair pulling, trichotillomania, and anxiety (Stewart, Jenike, & Keuthen, 2005). The complexity of OCD features made it challenging for treatment options. Most of the molecular studies in OCD are focused on genetic concept and genome-wide association studies of the disorder (Lin, Cao, & Gao, 2015; Zamanian-Azodi et al., 2015).
However, the pathophysiological origin of mental disorders, such as OCD, is still elusive despite these investigations. On the other hand, identification of the key relevant proteins in the disease condition is valuable for substantial insight of psychiatric disorders. By understanding etiological biomarkers of the disease, the underling mechanisms can be also explained (Filiou, Turck, & Martins‐de‐Souza, 2010). Body fluids are very useful sources to detect the biomarker; 1 of the appropriate ones is serum. Serum contains the vast range of proteins originated from normal or abnormal function of cells and tissue processes (Samavat et al., 2015; Tang, Beer, & Speicher, 2011). Moreover, serum is a suitable source (easily sampling and preparation) for different kinds of investigations as well as proteomics. Proteomic studies are important to determine expression levels of the proteins (Zali, Zamanian-Azodi, Tavirani, & Baghban, 2015). It is a powerful tool that provides useful information besides the other molecular studies (Rezaei-Tavirani et al., 2016). As mentioned earlier, OCD has various subtype models. One of the common models for the females is the washing compulsion (Noshirvani, Kasvikis, Marks, Tsakiris, & Monteiro, 1991). Proteins and mechanisms that correspond to the risk of this subtype are yet to be studied. To the authors` best knowledge, the current study was the 1st proteome analysis of obsessive-compulsive disorder. In the current pilot study, the proteome profile of patients with OCD washing phenotype was studied by 2D electrophoresis and bioinformatics.
2. Methods
2.1. Sample collection
2.1.1. Human subjects
The washing model cases (5 female patients) with moderate severity and control group (5 healthy volunteers) from Taleghani Hospital, Tehran, Iran, were diagnosed based on the diagnostic and statistical manual of mental disorders, 5th edition (DSM-5). The cases signed written informed consents, and the control and case groups were demographically matched. The proteome of these pooled medication-free patients was compared with that of the pooled controls.
2.1.2. Sample preparation
Blood collection was handled by venipuncture rout and 20-gauge needle. The samples were kept at room temperature for 30 minutes. After clotting, serum samples were separated by 2 times centrifugation at 2000g for 10 minutes at 4°C. Finally, the samples were maintained at −80°C for future processes (Nejadi et al., 2015). The protein extraction was conducted by the acetone precipitation method, according to introduction of Sigma ProteoPrep Protein Precipitation Kit.
2.1.3. Proteomic experiment
All 2DE chemicals and Ready Strip™ IPG strips were provided by GE Health Life Science. Prior to 2DE, the total protein concentration of the samples was determined by Bradford assay. The 2DE procedure was handled with 3 replications of normal and OCD samples. The 1st dimension, isoelectric focusing (IEF) was carried out by the application of Bio-Rad Protein IEF Cell, 7cm nonlinear IPG with the pH range of 3 to 11. In this step, about 500µg protein was loaded for each gel. The IEF separates proteins based on their pI. In the 2nd dimension, proteins were separated based on Molecular Weight (MW) by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and buffering systems in electrophoresis tank (Bio-Rad). After electrophoresis, the gels were dyed by Coomassie blue stating method and, then, scanned using Bio-Rad scanner (Hasanzadeh, Rezaie-Tavirani, Seyyedi, & Emadi, 2015). Finally, Progenesis SameSpots software was employed to analyze protein expression changes. A value of 1.5-fold increase or decrease was used as a cut off. Statistically significant differences (P≤0.05) in spot intensities were identified using 1-way ANOVA analysis. The significantly altered proteins in the expression were identified by http://world-2dpage.expasy.org/swiss-2dpage/. The amounts of MW and pI were the indices to identify proteins from this database.
2.2. Network analysis
Further investigation, based on interaction analysis, was carried out by Cytoscape 3.4.0-Milestone 2 software (Bader & Hogue, 2003). STRING database (DB) was the interaction source for PPI analysis by Cytoscape. STRING has the feature of providing comprehensive information from both experimental and predicted interactions of different databases with a probabilistic confidence score (Szklarczyk et al., 2010). Proteins identified by 2DE experiment were searched through STRING DB integrated in Cytoscape. The protein names were the query inputs and the selected species for this query was Homo sapiens as available in the query box.
The confidence (score) cut off for interactions was set to 0.5. Furthermore, about 100 additional nodes were added to the current study investigated proteins to expand the network. Network Analyzer plug-in analyzed functional topological parameters (centrality) including node degree and betweenness centrality (BC). This application is a powerful software to examine biological networks for the centrality parameters calculation by resourceful graph algorithms (Assenov, Ramírez, Schelhorn, Lengauer, & Albrecht, 2007).
Nodes that have high degree and betweenness values are the hub-bottleneck elements (Rezaei-Tavirani et al., 2016). MCODE, a Cytoscape Plug-in, examines protein complexes (regions with high interconnections) in PPI network. This Cytoscape plug-in is a clustering algorithm that examines the modules in a PPI network. Highly connected parts are detected by identifying nodes (seed) that are locally dense, travel outward from it, find the local neighbors, and isolate them as a cluster. The clusters are ranked based on the interconnection scores. Another level of functional annotations can be introduced by identifying the protein clusters. That is, proteins in a specific cluster usually indicate similar annotations (Bader & Hogue, 2003).
Finally, functional annotations of the current study investigated proteins and the detected modules were analyzed by the application of ClueGO plug-in (Bindea et al., 2009). GlueGO2.1.7 and its extended tool CluePedia annoted genes for biological process (BP). ClueGO presents gene ontology ranging from general to very specific ones as groups of related terms with similar shared proteins. The linkage strength between the terms, calculated by kappa score, was 0 to 1 (Bindea, Galon, & Mlecnik, 2013; Bindea et al., 2009). The kappa score was set to 0.5 for BP analysis. Minimum and maximum levels of ontology were set to 3 and 8 as the default option, respectively. The P was also set to ≤0.05. The correction method for P≤0.05 was Bonferroni step down method. The enrichment/depletion test for the terms was set to 2-sided enrichment/depletion, based on hypergeometric.
3. Results
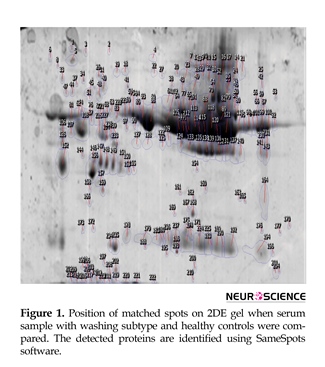
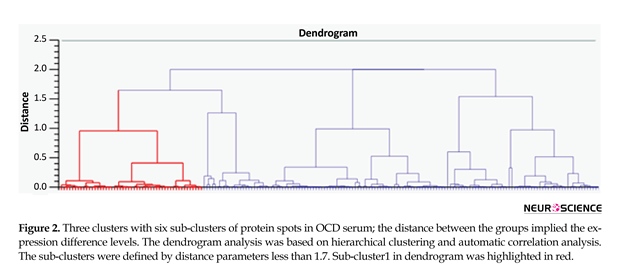
A total of 41 spots were differentially abundant in the OCD type that 18 showed upregulation and 23 downregulation. The abundant proteins, such as albumin and immunoglobulin, were not depleted in the current study. In Figure 1, positions of protein spots in this investigation are depicted. Progenesis SameSpots software revealed hierarchical clusters of proteins with similar expression correlations as dendrogram (Figure 2).
Obsessive-Compulsive Disorder (OCD) is a complex neuropsychological condition with a lifetime prevalence of 1% to 2.5% in the general population (Campos, Yoshimi, Simão, Torresan, & Torres, 2015). The World Health Organization (WHO) introduced OCD as a leading cause of non-fatal illness-related disability that primarily affects people with the age range of 15 to 44 years (Zilhão et al., 2015). This anxiety disorder is characterized by intrusive thoughts and compulsive behaviors (Carmi, Dar, Zohar, & Zangen, 2015). Dysfunction in corticostriatal circuits is the main known implicated part of brain in pathology of OCD and OC-spectrum disorders (Burguiere, Monteiro, Mallet, Feng, & Graybiel, 2015).
The complex nature of this heterogeneous disorder is described by the influence of molecular and environmental interactions (Jaffe et al., 2014). That is, the combination of many factors is responsible for manifestation of this disorder with different subtypes. The contribution of many genes with their polymorphisms expresses different subtypes with overlapped symptom dimensions. This complex clinical feature of OCD is accompanied by other related disorders (comorbidity), including Tourette syndrome, chronic hair pulling, trichotillomania, and anxiety (Stewart, Jenike, & Keuthen, 2005). The complexity of OCD features made it challenging for treatment options. Most of the molecular studies in OCD are focused on genetic concept and genome-wide association studies of the disorder (Lin, Cao, & Gao, 2015; Zamanian-Azodi et al., 2015).
However, the pathophysiological origin of mental disorders, such as OCD, is still elusive despite these investigations. On the other hand, identification of the key relevant proteins in the disease condition is valuable for substantial insight of psychiatric disorders. By understanding etiological biomarkers of the disease, the underling mechanisms can be also explained (Filiou, Turck, & Martins‐de‐Souza, 2010). Body fluids are very useful sources to detect the biomarker; 1 of the appropriate ones is serum. Serum contains the vast range of proteins originated from normal or abnormal function of cells and tissue processes (Samavat et al., 2015; Tang, Beer, & Speicher, 2011). Moreover, serum is a suitable source (easily sampling and preparation) for different kinds of investigations as well as proteomics. Proteomic studies are important to determine expression levels of the proteins (Zali, Zamanian-Azodi, Tavirani, & Baghban, 2015). It is a powerful tool that provides useful information besides the other molecular studies (Rezaei-Tavirani et al., 2016). As mentioned earlier, OCD has various subtype models. One of the common models for the females is the washing compulsion (Noshirvani, Kasvikis, Marks, Tsakiris, & Monteiro, 1991). Proteins and mechanisms that correspond to the risk of this subtype are yet to be studied. To the authors` best knowledge, the current study was the 1st proteome analysis of obsessive-compulsive disorder. In the current pilot study, the proteome profile of patients with OCD washing phenotype was studied by 2D electrophoresis and bioinformatics.
2. Methods
2.1. Sample collection
2.1.1. Human subjects
The washing model cases (5 female patients) with moderate severity and control group (5 healthy volunteers) from Taleghani Hospital, Tehran, Iran, were diagnosed based on the diagnostic and statistical manual of mental disorders, 5th edition (DSM-5). The cases signed written informed consents, and the control and case groups were demographically matched. The proteome of these pooled medication-free patients was compared with that of the pooled controls.
2.1.2. Sample preparation
Blood collection was handled by venipuncture rout and 20-gauge needle. The samples were kept at room temperature for 30 minutes. After clotting, serum samples were separated by 2 times centrifugation at 2000g for 10 minutes at 4°C. Finally, the samples were maintained at −80°C for future processes (Nejadi et al., 2015). The protein extraction was conducted by the acetone precipitation method, according to introduction of Sigma ProteoPrep Protein Precipitation Kit.
2.1.3. Proteomic experiment
All 2DE chemicals and Ready Strip™ IPG strips were provided by GE Health Life Science. Prior to 2DE, the total protein concentration of the samples was determined by Bradford assay. The 2DE procedure was handled with 3 replications of normal and OCD samples. The 1st dimension, isoelectric focusing (IEF) was carried out by the application of Bio-Rad Protein IEF Cell, 7cm nonlinear IPG with the pH range of 3 to 11. In this step, about 500µg protein was loaded for each gel. The IEF separates proteins based on their pI. In the 2nd dimension, proteins were separated based on Molecular Weight (MW) by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and buffering systems in electrophoresis tank (Bio-Rad). After electrophoresis, the gels were dyed by Coomassie blue stating method and, then, scanned using Bio-Rad scanner (Hasanzadeh, Rezaie-Tavirani, Seyyedi, & Emadi, 2015). Finally, Progenesis SameSpots software was employed to analyze protein expression changes. A value of 1.5-fold increase or decrease was used as a cut off. Statistically significant differences (P≤0.05) in spot intensities were identified using 1-way ANOVA analysis. The significantly altered proteins in the expression were identified by http://world-2dpage.expasy.org/swiss-2dpage/. The amounts of MW and pI were the indices to identify proteins from this database.
2.2. Network analysis
Further investigation, based on interaction analysis, was carried out by Cytoscape 3.4.0-Milestone 2 software (Bader & Hogue, 2003). STRING database (DB) was the interaction source for PPI analysis by Cytoscape. STRING has the feature of providing comprehensive information from both experimental and predicted interactions of different databases with a probabilistic confidence score (Szklarczyk et al., 2010). Proteins identified by 2DE experiment were searched through STRING DB integrated in Cytoscape. The protein names were the query inputs and the selected species for this query was Homo sapiens as available in the query box.
The confidence (score) cut off for interactions was set to 0.5. Furthermore, about 100 additional nodes were added to the current study investigated proteins to expand the network. Network Analyzer plug-in analyzed functional topological parameters (centrality) including node degree and betweenness centrality (BC). This application is a powerful software to examine biological networks for the centrality parameters calculation by resourceful graph algorithms (Assenov, Ramírez, Schelhorn, Lengauer, & Albrecht, 2007).
Nodes that have high degree and betweenness values are the hub-bottleneck elements (Rezaei-Tavirani et al., 2016). MCODE, a Cytoscape Plug-in, examines protein complexes (regions with high interconnections) in PPI network. This Cytoscape plug-in is a clustering algorithm that examines the modules in a PPI network. Highly connected parts are detected by identifying nodes (seed) that are locally dense, travel outward from it, find the local neighbors, and isolate them as a cluster. The clusters are ranked based on the interconnection scores. Another level of functional annotations can be introduced by identifying the protein clusters. That is, proteins in a specific cluster usually indicate similar annotations (Bader & Hogue, 2003).
Finally, functional annotations of the current study investigated proteins and the detected modules were analyzed by the application of ClueGO plug-in (Bindea et al., 2009). GlueGO2.1.7 and its extended tool CluePedia annoted genes for biological process (BP). ClueGO presents gene ontology ranging from general to very specific ones as groups of related terms with similar shared proteins. The linkage strength between the terms, calculated by kappa score, was 0 to 1 (Bindea, Galon, & Mlecnik, 2013; Bindea et al., 2009). The kappa score was set to 0.5 for BP analysis. Minimum and maximum levels of ontology were set to 3 and 8 as the default option, respectively. The P was also set to ≤0.05. The correction method for P≤0.05 was Bonferroni step down method. The enrichment/depletion test for the terms was set to 2-sided enrichment/depletion, based on hypergeometric.
3. Results
A total of 41 spots were differentially abundant in the OCD type that 18 showed upregulation and 23 downregulation. The abundant proteins, such as albumin and immunoglobulin, were not depleted in the current study. In Figure 1, positions of protein spots in this investigation are depicted. Progenesis SameSpots software revealed hierarchical clusters of proteins with similar expression correlations as dendrogram (Figure 2).
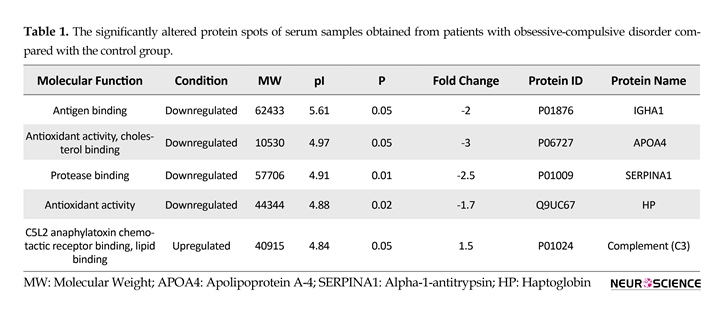
Five identified proteins with their properties are tabulated in Table 1. The codes and molecular functions of the elements were retrieved from Uniprot database. The position, MW and pI, and the condition of regulation of each specific protein in the samples obtained from patients with were found through the proteomic analysis.
The proteins were identified by http://world-2dpage.expasy.org/swiss-2dpage/. The amounts of MW and pI were the indices to identify proteins from this database. SameSpots software analyzed the position, MW and pI, and condition of regulation (up- or downregulation) of each specific protein in the samples of patients with OCD.
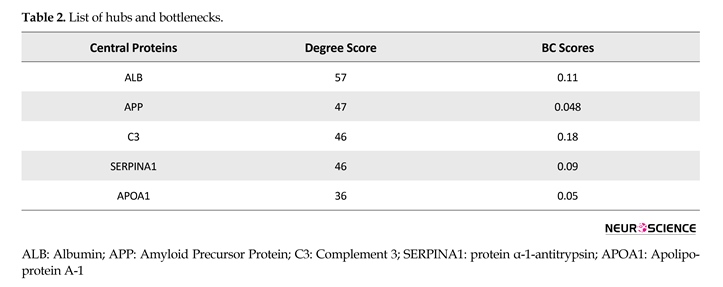
PPI networks provide noteworthy information related to mechanisms of a specific disease (Safaei et al., 2016). Only 4 out of 5 proteins were involved in the current study PPI network. The interaction network was visualized by Cytoscape software version 3.4.0-Milestone 2 (Figure 3). Central properties of PPI network were analyzed using Network Analyzer application. Proteins with high degree and betweenness centrality scores were selected and presented (Table 2).
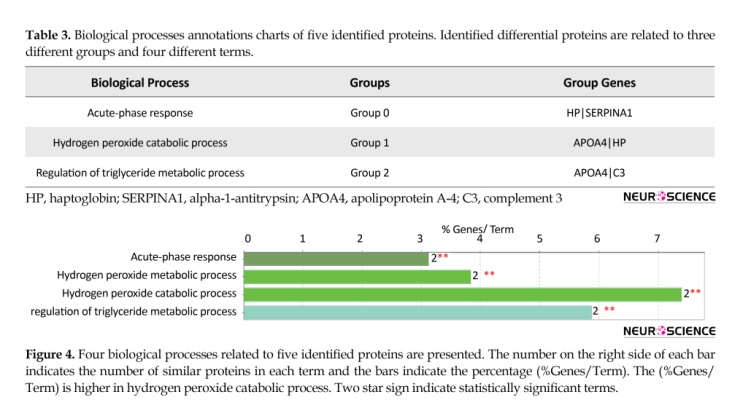
The proteins were ranked based on degree scores. Five central nodes were tabulated, out of which albumin showed the highest value. The C3 and SERPINA1 were the identified proteins in OCD gel sample and the 3 other proteins were imported from the network nodes. To determine the BP annotations of the selected proteins and investigated modules, ClueGO application was used to provide a better understanding of OCD pathophysiological processes (Table 3 and Figure 4).
The identified proteins were related to 3 different groups and 4 different terms. The number on the right side of each bar indicates the number of similar proteins in each term and the bars indicate the percentage (%Genes/Term). The (%Genes/Term) was higher in hydrogen peroxide catabolic process. Two-star sign is expressed the statistically significant values. The PPI network contained complex protein structures of densely
The proteins were identified by http://world-2dpage.expasy.org/swiss-2dpage/. The amounts of MW and pI were the indices to identify proteins from this database. SameSpots software analyzed the position, MW and pI, and condition of regulation (up- or downregulation) of each specific protein in the samples of patients with OCD.
PPI networks provide noteworthy information related to mechanisms of a specific disease (Safaei et al., 2016). Only 4 out of 5 proteins were involved in the current study PPI network. The interaction network was visualized by Cytoscape software version 3.4.0-Milestone 2 (Figure 3). Central properties of PPI network were analyzed using Network Analyzer application. Proteins with high degree and betweenness centrality scores were selected and presented (Table 2).
The proteins were ranked based on degree scores. Five central nodes were tabulated, out of which albumin showed the highest value. The C3 and SERPINA1 were the identified proteins in OCD gel sample and the 3 other proteins were imported from the network nodes. To determine the BP annotations of the selected proteins and investigated modules, ClueGO application was used to provide a better understanding of OCD pathophysiological processes (Table 3 and Figure 4).
The identified proteins were related to 3 different groups and 4 different terms. The number on the right side of each bar indicates the number of similar proteins in each term and the bars indicate the percentage (%Genes/Term). The (%Genes/Term) was higher in hydrogen peroxide catabolic process. Two-star sign is expressed the statistically significant values. The PPI network contained complex protein structures of densely
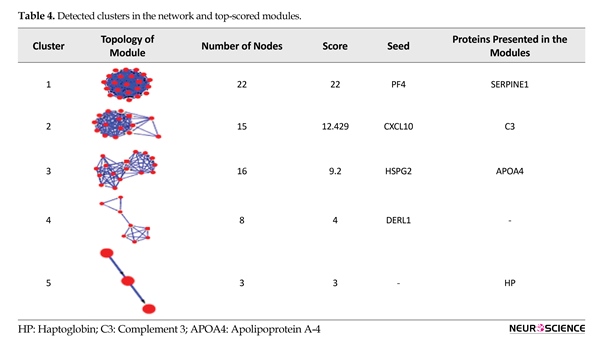
connected regions. Protein complexes of OCD PPI network were retrieved by MCODE (Table 4).
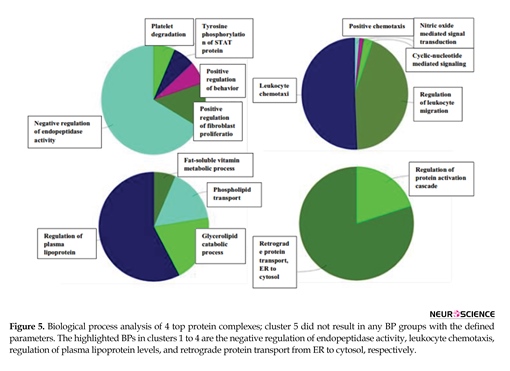
Eight clusters were detected in the network and 5 top-scored modules are presented. Cluster members are shown in red, and square nodes represent the seeds. Seed proteins are the important nodes with highest interaction score in the module. Seed proteins are PF4, CXCL10, HSPG2, and DERL1 for the clusters 1 to 4, respectively. SERPINE1, C3, APOA4, and HP are also present in the clusters 1 to 5, respectively. The statistical parameters of the analysis was based on the cutoff point of 2 and node score cutoff point of 0.2. Biological process analysis of four top clusters is presented in Figure 5.
Eight clusters were detected in the network and 5 top-scored modules are presented. Cluster members are shown in red, and square nodes represent the seeds. Seed proteins are the important nodes with highest interaction score in the module. Seed proteins are PF4, CXCL10, HSPG2, and DERL1 for the clusters 1 to 4, respectively. SERPINE1, C3, APOA4, and HP are also present in the clusters 1 to 5, respectively. The statistical parameters of the analysis was based on the cutoff point of 2 and node score cutoff point of 0.2. Biological process analysis of four top clusters is presented in Figure 5.
4. Discussion
Molecular evaluation of obsessive-compulsive disorder is important due to the complex nature of the disorder. This feature resulted in an increased interest in the genetic studies. That is, most of the related molecular studies are primarily focused on genetic and genome concepts (Zamanian-Azodi et al., 2015). In this sense, researches show that many genes with related polymorphisms play key roles in the pathogenesis of OCD (Den Braber et al., 2016). On the other hand, proteomic study of OCD is not investigated yet. Proteomics can help to understand proteome changes of human during the disease state. In fact, many potential proteins may have expression alterations in this condition. The specific changed proteins are known as possible biomarkers that can facilitate diagnosis and treatment approaches in anxiety disorders. The current study analyzed proteins with significant expression changes in response to OCD pathogenesis. As shown in Figure 1, some protein spots in proteome profile of patient with OCD can have expression alterations. The presence of highly abundant proteins can have 2 opposite features.
Molecular evaluation of obsessive-compulsive disorder is important due to the complex nature of the disorder. This feature resulted in an increased interest in the genetic studies. That is, most of the related molecular studies are primarily focused on genetic and genome concepts (Zamanian-Azodi et al., 2015). In this sense, researches show that many genes with related polymorphisms play key roles in the pathogenesis of OCD (Den Braber et al., 2016). On the other hand, proteomic study of OCD is not investigated yet. Proteomics can help to understand proteome changes of human during the disease state. In fact, many potential proteins may have expression alterations in this condition. The specific changed proteins are known as possible biomarkers that can facilitate diagnosis and treatment approaches in anxiety disorders. The current study analyzed proteins with significant expression changes in response to OCD pathogenesis. As shown in Figure 1, some protein spots in proteome profile of patient with OCD can have expression alterations. The presence of highly abundant proteins can have 2 opposite features.
While it is possible to discover the feasible expression changes in these proteins, they can mask the detection of low abundance proteins (Nejadi et al., 2015).
In the current preliminary experiment, 1 of the main goals was to detect high abundant profile changes as well. Figure 2 indicates the clustering profile of all present proteins in OCD gel. Three clusters and 6 sub-clusters based on expression changes were obtained. The 1st sub-cluster is highlighted in red and the spots in each sub-cluster have close correlation values. The smaller the protein clusters, the bigger the correlation values. Proteins in each sub-cluster, especially those within smaller clusters, may show common ontology properties as their pattern of expression changes are similar. Detecting differentially expressed abundant serum proteins in OCD may provide information of other mechanisms of disorder development.
In most of the proteomic studies, these proteins are normally subjected to specific methods of depletion (Corbett et al., 2006). The possibility of contribution of these proteins in similar processes may be higher than that of the low-abundance proteins in serum. The reason for this claim is that low-abundance proteins are mostly secreted to the serum form different parts of the body and may be related to different pathways, while the main resident proteins of serum may be more correlated in functional properties. Observed proteins in the proteomic profile of patients with OCD, as indicated in Table 1, are IGHA1, APOA4, HP, SERPINA1, and C3.
There were significantly different expression patterns in the comparison of normal and patient samples. The first 4 proteins showed down-regulation, while C3 showed upregulation in the patient samples. Therefore, these proteins may have a central role in the disorder manifestation. Four of the proteins belonged to the 6 most abundant proteins in the serum including IgA, haptoglobin (HP), transferrin, and antitrypsin. Immunoglobulin heavy constant alpha (IGHA1) is a major immunoglobulin, here, the serum level of this protein changes significantly in the patients with OCD; however, it is not reported as a candidate protein in other mental disorders. APOA4, as a low abundant protein, belongs to the apolipoprotein superfamily. Apolipoproteins regulate the level of free fatty acids in plasma, high-density lipoprotein, and triglyceride-rich lipoproteins metabolisms, and have a role in the reverse cholesterol transport pathway (Levin et al., 2009).
In the current preliminary experiment, 1 of the main goals was to detect high abundant profile changes as well. Figure 2 indicates the clustering profile of all present proteins in OCD gel. Three clusters and 6 sub-clusters based on expression changes were obtained. The 1st sub-cluster is highlighted in red and the spots in each sub-cluster have close correlation values. The smaller the protein clusters, the bigger the correlation values. Proteins in each sub-cluster, especially those within smaller clusters, may show common ontology properties as their pattern of expression changes are similar. Detecting differentially expressed abundant serum proteins in OCD may provide information of other mechanisms of disorder development.
In most of the proteomic studies, these proteins are normally subjected to specific methods of depletion (Corbett et al., 2006). The possibility of contribution of these proteins in similar processes may be higher than that of the low-abundance proteins in serum. The reason for this claim is that low-abundance proteins are mostly secreted to the serum form different parts of the body and may be related to different pathways, while the main resident proteins of serum may be more correlated in functional properties. Observed proteins in the proteomic profile of patients with OCD, as indicated in Table 1, are IGHA1, APOA4, HP, SERPINA1, and C3.
There were significantly different expression patterns in the comparison of normal and patient samples. The first 4 proteins showed down-regulation, while C3 showed upregulation in the patient samples. Therefore, these proteins may have a central role in the disorder manifestation. Four of the proteins belonged to the 6 most abundant proteins in the serum including IgA, haptoglobin (HP), transferrin, and antitrypsin. Immunoglobulin heavy constant alpha (IGHA1) is a major immunoglobulin, here, the serum level of this protein changes significantly in the patients with OCD; however, it is not reported as a candidate protein in other mental disorders. APOA4, as a low abundant protein, belongs to the apolipoprotein superfamily. Apolipoproteins regulate the level of free fatty acids in plasma, high-density lipoprotein, and triglyceride-rich lipoproteins metabolisms, and have a role in the reverse cholesterol transport pathway (Levin et al., 2009).
Low expression of APOA4 may influence cholesterol level reduction in the serum of patients with OCD. This phenomenon may lead to dysregulation of neurons neurochemical (hyperactivity of serotonin reuptake receptor activity), as cholesterol is the main component of neurons (Madhura, 2015). Another function of APOA4 is as an antioxidant agent (Qin, Swertfeger, Zheng, Hui, & Tso, 1998); this role may justify the accumulation of oxidative product during OCD pathophysiology (Şimşek, Gençoğlan, & Yüksel, 2016). The downregulation of this protein is also reported in schizophrenia and the Alzheimer Disease (AD) (Levin et al., 2009; Lin, Cao, & Gao, 2015). HP is another serum abundant protein that binds to hemoglobin and has chaperone function. This protein shows downregulation in AD (Cocciolo et al., 2012).
Protein α-1-antitrypsin (SERPINA1) is the most abundant protease inhibitor in serum release from liver (Pastore, Ballabio, & Brunetti-Pierri, 2013). It shows down-regulation in OCD profile. This protein is also counted as a candidate biomarker in pathophysiology of schizophrenia (Davalieva, Kostovska, & Dwork, 2016). Complement C3 as a part of complement system is responsible for innate defense mechanism against pathogenic microorganisms (Ehrnthaller, Ignatius, Gebhard, & Huber-Lang, 2011). Increased level of C3 in the patients with OCD was observed in the current study. It may indicate that, immunity-related pathways are as part of the OCD underlying mechanism. Similar to neurodegenerative disorders, maintained activation of inflammation level in brain implies the protective effects to reduce toxic products of these brain disorders (Bonifati & Kishore, 2007).
In patients with schizophrenia, increased level of C3 level is also reported (Mayilyan, Weinberger, & Sim, 2008). Generation of oxidative products can be related to exaggerated or insufficient activation of the complement system (Bonifati & Kishore, 2007). Increment of oxidative products is reported in OCD (Şimşek et al., 2016). In fact, there are some overlaps between mental disorders and biomarkers of other neurological diseases. As mentioned above, expression alterations of APOA4, HP, and C3 proteins in OCD profile were also mentioned in other neurological diseases (Davalieva et al., 2016). Investigation of these proteins through PPI network can provide a better understanding of the prominent role of the detected proteins. The biological functions of identified proteins can be evaluated in this way (Levin et al., 2009). The expression changes of these proteins in serum may indicate integrative linkage of inflammatory response systems and the risk of OCD as it is also approved in other mental disorders such as schizophrenia (Davalieva et al., 2016).
In Figure 3, direct link among APOA4, HP, and SERPINA1 may specify an interaction among them. Network topology analysis showed central features of 2 proteins of C3 and SERPINA1 in the network as tabulated in Table 2. This analysis detected 3 proteins (ALB, APP, and APOA1) as candidates for central proteins that may be related to OCD risk. These proteins are also important in disorders such as schizophrenia (Huang, 2002; Levin et al., 2009; Yang et al., 2006). Biological processes related to differentially changed proteins may be remarkably influenced. As mentioned earlier, 1 of the important changes in brain diseases is the accumulation of oxidative products. This fact is also confirmed by gene ontology analysis of proteins identified in OCD shown in Table 3.
Hydrogen peroxide catabolic process is the primary-linked annotation of the identified proteins. The dysregulation of it may be one of the reasons of presentation of oxidative products. Further investigation of network topology identified 8 complexes of proteins, that the identified proteins in the current study are present in 4 of the top ranked ones. As shown in Table 4, central proteins in the network, SERPINA1 and C3, also belong to the 1st top clusters. This fact may imply the additionally important role of these proteins in the network integrity. The 5 top ranked protein complexes were further analyzed for biological process enrichment as depicted in Figure 5. The significant processes for the cluster 1 to 4 were the negative regulation of endopeptidase activity, leukocyte chemotaxis, regulation of plasma lipoprotein levels, and retrograde protein transport, ER to cytosol, respectively. These annotations may have important roles in OCD related mechanisms.
Finally, proteomic analysis and proteins identified in PPI network construction and their related processes were previously reported in other brain diseases as well. Consequently, literature review can be counted as an approach to validate the current preliminary study. Overall, the findings may be useful to understand molecular behavior of OCD and provide a starting point for further investigations of OCD proteome profile changes.
In conclusion, proteins involved in the OCD risk, provide new insight on the complexity of the disorder. IGHA1, APOA4, HP, SERPINA1, and C3 with significant expression changes, in particular the last 2 mentioned proteins with high centrality properties, may serve as potential treatment targets; however, furthered research is required to validate the preliminary information. It is also suggested to improve OCD diagnosis and treatment approaches by analyzing serum proteome with the focus on low-abundance proteins.
Protein α-1-antitrypsin (SERPINA1) is the most abundant protease inhibitor in serum release from liver (Pastore, Ballabio, & Brunetti-Pierri, 2013). It shows down-regulation in OCD profile. This protein is also counted as a candidate biomarker in pathophysiology of schizophrenia (Davalieva, Kostovska, & Dwork, 2016). Complement C3 as a part of complement system is responsible for innate defense mechanism against pathogenic microorganisms (Ehrnthaller, Ignatius, Gebhard, & Huber-Lang, 2011). Increased level of C3 in the patients with OCD was observed in the current study. It may indicate that, immunity-related pathways are as part of the OCD underlying mechanism. Similar to neurodegenerative disorders, maintained activation of inflammation level in brain implies the protective effects to reduce toxic products of these brain disorders (Bonifati & Kishore, 2007).
In patients with schizophrenia, increased level of C3 level is also reported (Mayilyan, Weinberger, & Sim, 2008). Generation of oxidative products can be related to exaggerated or insufficient activation of the complement system (Bonifati & Kishore, 2007). Increment of oxidative products is reported in OCD (Şimşek et al., 2016). In fact, there are some overlaps between mental disorders and biomarkers of other neurological diseases. As mentioned above, expression alterations of APOA4, HP, and C3 proteins in OCD profile were also mentioned in other neurological diseases (Davalieva et al., 2016). Investigation of these proteins through PPI network can provide a better understanding of the prominent role of the detected proteins. The biological functions of identified proteins can be evaluated in this way (Levin et al., 2009). The expression changes of these proteins in serum may indicate integrative linkage of inflammatory response systems and the risk of OCD as it is also approved in other mental disorders such as schizophrenia (Davalieva et al., 2016).
In Figure 3, direct link among APOA4, HP, and SERPINA1 may specify an interaction among them. Network topology analysis showed central features of 2 proteins of C3 and SERPINA1 in the network as tabulated in Table 2. This analysis detected 3 proteins (ALB, APP, and APOA1) as candidates for central proteins that may be related to OCD risk. These proteins are also important in disorders such as schizophrenia (Huang, 2002; Levin et al., 2009; Yang et al., 2006). Biological processes related to differentially changed proteins may be remarkably influenced. As mentioned earlier, 1 of the important changes in brain diseases is the accumulation of oxidative products. This fact is also confirmed by gene ontology analysis of proteins identified in OCD shown in Table 3.
Hydrogen peroxide catabolic process is the primary-linked annotation of the identified proteins. The dysregulation of it may be one of the reasons of presentation of oxidative products. Further investigation of network topology identified 8 complexes of proteins, that the identified proteins in the current study are present in 4 of the top ranked ones. As shown in Table 4, central proteins in the network, SERPINA1 and C3, also belong to the 1st top clusters. This fact may imply the additionally important role of these proteins in the network integrity. The 5 top ranked protein complexes were further analyzed for biological process enrichment as depicted in Figure 5. The significant processes for the cluster 1 to 4 were the negative regulation of endopeptidase activity, leukocyte chemotaxis, regulation of plasma lipoprotein levels, and retrograde protein transport, ER to cytosol, respectively. These annotations may have important roles in OCD related mechanisms.
Finally, proteomic analysis and proteins identified in PPI network construction and their related processes were previously reported in other brain diseases as well. Consequently, literature review can be counted as an approach to validate the current preliminary study. Overall, the findings may be useful to understand molecular behavior of OCD and provide a starting point for further investigations of OCD proteome profile changes.
In conclusion, proteins involved in the OCD risk, provide new insight on the complexity of the disorder. IGHA1, APOA4, HP, SERPINA1, and C3 with significant expression changes, in particular the last 2 mentioned proteins with high centrality properties, may serve as potential treatment targets; however, furthered research is required to validate the preliminary information. It is also suggested to improve OCD diagnosis and treatment approaches by analyzing serum proteome with the focus on low-abundance proteins.
Acknowledgements
Authors would like to thank Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, for the support. The current study was derived from the PhD thesis of Miss Mona Zamanian-Azodi.
Conflict of Interest
The authors declared no conflicts of interest.
References
Assenov, Y., Ramírez, F., Schelhorn, S. E., Lengauer, T., & Albrecht, M. (2007). Computing topological parameters of biological networks. Bioinformatics, 24(2), 282–284. doi: 10.1093/bioinformatics/btm554
Bader, G. D., & Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2. doi :10.1186/1471-2105-4-2
Bindea, G., Galon, J., & Mlecnik, B. (2013). CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics, 29(5), 661–3. doi: 10.1093/bioinformatics/btt019
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091–3. doi: 10.1093/bioinformatics/btp101
Bonifati, D., & Kishore, U. (2007). Role of complement in neurodegeneration and neuroinflammation. Molecular Immunology, 44(5), 999–1010. doi: 10.1016/j.molimm.2006.03.007
Burguière, E., Monteiro, P., Mallet, L., Feng, G., & Graybiel, A. M. (2015). Striatal circuits, habits, and implications for obsessive–compulsive disorder. Current Opinion in Neurobiology, 30, 59–65. doi: 10.1016/j.conb.2014.08.008
Campos, L. M., Yoshimi, N. T., Simão, M. O., Torresan, R. C., & Torres, A. R. (2015). Obsessive-compulsive symptoms among alcoholics in outpatient treatment: Prevalence, severity and correlates. Psychiatry Research, 229(1-2), 401–9. doi: 10.1016/j.psychres.2015.05.111
Carmi, L., Al Yagon U., Dar, R., Zohar, J., & Zangen, A. (2015). Deep Transcranial Magnetic Stimulation (TMS) in Obsessive Compulsive Disorder (OCD) patients. European Psychiatry, 30, 794. doi: 10.1016/s0924-9338(15)30618-0
Cocciolo, A., di Domenico, F., Coccia, R., Fiorini, A., Cai, J., Pierce, W. M., et al. (2012). Decreased expression and increased oxidation of plasma haptoglobin in Alzheimer disease: Insights from redox proteomics. Free Radical Biology and Medicine, 53(10), 1868–76. doi: 10.1016/j.freeradbiomed.2012.08.596
Corbett, B. A., Kantor, A. B., Schulman, H., Walker, W. L., Lit, L., Ashwood, P., et al. (2006). A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Molecular Psychiatry, 12(3), 292-306. doi: 10.1038/sj.mp.4001943
Davalieva, K., Maleva Kostovska, I., & Dwork, A. J. (2016). Proteomics research in schizophrenia. Frontiers in Cellular Neuroscience, 10, 18. doi: 10.3389/fncel.2016.00018
Den Braber, A., Zilhão, N. R., Fedko, I. O., Hottenga, J. J., Pool, R., Smit, D. J. A., et al. (2016). Obsessive–compulsive symptoms in a large population-based twin-family sample are predicted by clinically based polygenic scores and by genome-wide SNPs. Translational Psychiatry, 6(2), 731. doi: 10.1038/tp.2015.223
Ehrnthaller, C., Ignatius, A., Gebhard, F., & Huber-Lang, M. (2011). New insights of an old defense system: Structure, function, and clinical relevance of the complement system. Molecular Medicine, 17(3), 317-29. PMID: 21046060
Filiou, M. D., Turck, C. W., & Martins-de-Souza, D. (2010). Quantitative proteomics for investigating psychiatric disorders. PROTEOMICS - Clinical Applications, 5(1-2), 38–49. doi: 10.1002/prca.201000060
Hasanzadeh, H., Rezaie-Tavirani, M., Seyyedi, S., & Emadi, A. (2015). [Proteomics study of extremely low frequency electromagnetic field (50 Hz) on human neuroblastoma cells (Persian)]. Koomesh, 17(1), 233-239.
Huang, T. L. (2002). Decreased serum albumin levels in Taiwanese patients with schizophrenia. Psychiatry and Clinical Neurosciences, 56(6), 627–30. doi: 10.1046/j.1440-1819.2002.01066.x
Jaffe, A. E., Deep-Soboslay, A., Tao, R., Hauptman, D. T., Kaye, W. H., Arango, V., et al. (2014). Genetic neuropathology of obsessive psychiatric syndromes. Translational Psychiatry, 4(9), 432. doi: 10.1038/tp.2014.68
Levin, Y., Wang, L., Schwarz, E., Koethe, D., Leweke, F. M., & Bahn, S. (2009). Global proteomic profiling reveals altered proteomic signature in schizophrenia serum. Molecular Psychiatry, 15(11), 1088–100. doi: 10.1038/mp.2009.54
Liu, S., Cui, J., Wang, X., Ma, J., Niu, H., Ma, X., et al. (2015). An association study between DLGAP1 rs11081062 and EFNA5 rs26728 polymorphisms with obsessive–compulsive disorder in a Chinese Han population. Neuropsychiatric Disease and Treatment, 11, 897-905. doi: 10.2147/ndt.s75009
Lin, Q., Cao, Y., & Gao, J. (2015).Decreased expression of the APOA1-APOC3-APOA4 gene cluster is associated with risk of Alzheimer's disease. Drug Design, Development and Therapy, 9, 5421-31. doi: 10.2147/dddt.s89279
Madhura, T. K. (2015). Role of serum cholesterol level in pathogenesis of OCD. Biochemistry & Analytical Biochemistry, 4(3), 1000183. doi: 10.4172/2161-1009.1000183
Mayilyan, K. R., Weinberger, D. R., & Sim, R. B. (2008). The complement system in schizophrenia. Drug News & Perspectives, 21(4), 200-10. doi: 10.1358/dnp.2008.21.4.1213349
Nejadi, N., Hantoushzadeh, S., Masti Mohammadpoor, S., Hosseini, Z. S., Mousavi, M., & Gorgani-Firuzjaee, S. (2015). Vitamin D Binding Protein as screening biomarker candidate for late-onset preeclampsia without intrauterine growth restriction during 16 week of gestation. Journal of Paramedical Sciences, 6(1), 36-43.
Noshirvani, H. F., Kasvikis, Y., Marks, I. M., Tsakiris, F., & Monteiro, W. O. (1991). Gender-divergent aetiological factors in obsessive-compulsive disorder. The British Journal of Psychiatry, 158(2), 260–263. doi: 10.1192/bjp.158.2.260
Pastore, N., Ballabio, A., & Brunetti-Pierri, N. (2013). Autophagy master regulator TFEB induces clearance of toxic SERPINA1/α-1-antitrypsin polymers. Autophagy, 9(7), 1094–6. doi: 10.4161/auto.24469
Qin, X., Swertfeger, D. K., Zheng, S., Hui, D. Y., & Tso, P. (1998). Apolipoprotein AIV: A potent endogenous inhibitor of lipid oxidation. American Journal of Physiology-Heart and Circulatory Physiology, 274(5), 1836-40. PMID: 9612397
Zamanian-Azodi, M., Rezaei-Tavirani, M., Rahmati-Rad, S., & Rezaei Tavirani, M. (2016). Ethanol and cancer induce similar changes on protein expression pattern of human fibroblast cell. Iranian Journal of Pharmaceutical Research, 15, 175-84. PMID: 28228815
Rezaei-Tavirani, M., Zamanian-Azodi, M., Rajabi, S., Masoudi-Nejad, A., Rostami-Nejad, M., & Rahmatirad, S. (2016). Protein Clustering and Interactome Analysis in Parkinson and Alzheimer’s Diseases. Archives of Iranian Medicine, 19(2), 101-9. doi: 0161902/AIM.007
Safaei, A., Rezaei Tavirani, M., Arefi Oskouei, A., Zamanian Azodi, M., Mohebbi, S. R., & Nikzamir, A. R. (2016). Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterology and Hepatology From Bed to Bench, 9(2), 114-23. PMID: 27099671
Samavat, S., Kalantari, S., Nafar, M., Rutishauser, D., Rezaei-Tavirani, M., Parvin, M., et al. (2015). Diagnostic urinary proteome profile for immunoglobulin a nephropathy. Iranian Journal of Kidney Diseases, 9(3), 239-48. PMID: 25957429
Şimşek, Ş., Gençoğlan, S., & Yüksel, T. (2016). DNA damage and antioxidants in treatment naïve children with obsessive–compulsive disorder. Psychiatry Research, 237, 133–7. doi: 10.1016/j.psychres.2016.01.054
Stewart, S. E., Jenike, M. A., & Keuthen, N. J. (2005). Severe obsessive-compulsive disorder with and without comorbid hair pulling. Journal of Clinical Psychiatry, 66(7), 864–9. doi: 10.4088/jcp.v66n0709
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth A., Minguez, P., et al. (2010). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research, 39, 561-8. PMID: 21045058
Tang, H. Y., Beer, L. A., & Speicher, D. W. (2011). In-Depth Analysis of a Plasma or Serum Proteome Using a 4D Protein Profiling Method. Serum/Plasma Proteomics, 728, 47–67. doi: 10.1007/978-1-61779-068-3_3
Yang, Y., Wan, C., Li, H., Zhu, H., La, Y., Xi, Z., et al. (2006). Altered levels of acute phase proteins in the plasma of patients with schizophrenia. Analytical Chemistry, 78(11), 3571–6. doi: 10.1021/ac051916x
Zali, H., Zamanian-Azodi, M., Rezaei Tavirani, M., & Akbar-Zadeh Baghban, A. (2015). Protein drug targets of lavandula angustifolia on treatment of rat Alzheimer’s disease. Iranian Journal of Pharmaceutical Research, 14(1), 291-302. PMID: 25561935
Zamanian-Azodi, M., Rezaei-Tavirani, M., Kermani-Ranjbar, T., Oskouie, A. A, RahmatiRad, S., & Asl, N. S. (2015). [Pathophysiology, genetics, types, and treatments in obsessive compulsive disorder (Persian)]. Koomesh, 16(4), 475-87.
Zilhão, N. R., Smit, D. J. A., den Braber, A., Dolan, C. V., Willemsen, G., Boomsma, D. I., & Cath, D. C. (2014). Genetic and environmental contributions to stability in adult obsessive compulsive behavior. Twin Research and Human Genetics, 18(1), 52–60. doi: 10.1017/thg.2014.77
Authors would like to thank Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, for the support. The current study was derived from the PhD thesis of Miss Mona Zamanian-Azodi.
Conflict of Interest
The authors declared no conflicts of interest.
References
Assenov, Y., Ramírez, F., Schelhorn, S. E., Lengauer, T., & Albrecht, M. (2007). Computing topological parameters of biological networks. Bioinformatics, 24(2), 282–284. doi: 10.1093/bioinformatics/btm554
Bader, G. D., & Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2. doi :10.1186/1471-2105-4-2
Bindea, G., Galon, J., & Mlecnik, B. (2013). CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics, 29(5), 661–3. doi: 10.1093/bioinformatics/btt019
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091–3. doi: 10.1093/bioinformatics/btp101
Bonifati, D., & Kishore, U. (2007). Role of complement in neurodegeneration and neuroinflammation. Molecular Immunology, 44(5), 999–1010. doi: 10.1016/j.molimm.2006.03.007
Burguière, E., Monteiro, P., Mallet, L., Feng, G., & Graybiel, A. M. (2015). Striatal circuits, habits, and implications for obsessive–compulsive disorder. Current Opinion in Neurobiology, 30, 59–65. doi: 10.1016/j.conb.2014.08.008
Campos, L. M., Yoshimi, N. T., Simão, M. O., Torresan, R. C., & Torres, A. R. (2015). Obsessive-compulsive symptoms among alcoholics in outpatient treatment: Prevalence, severity and correlates. Psychiatry Research, 229(1-2), 401–9. doi: 10.1016/j.psychres.2015.05.111
Carmi, L., Al Yagon U., Dar, R., Zohar, J., & Zangen, A. (2015). Deep Transcranial Magnetic Stimulation (TMS) in Obsessive Compulsive Disorder (OCD) patients. European Psychiatry, 30, 794. doi: 10.1016/s0924-9338(15)30618-0
Cocciolo, A., di Domenico, F., Coccia, R., Fiorini, A., Cai, J., Pierce, W. M., et al. (2012). Decreased expression and increased oxidation of plasma haptoglobin in Alzheimer disease: Insights from redox proteomics. Free Radical Biology and Medicine, 53(10), 1868–76. doi: 10.1016/j.freeradbiomed.2012.08.596
Corbett, B. A., Kantor, A. B., Schulman, H., Walker, W. L., Lit, L., Ashwood, P., et al. (2006). A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Molecular Psychiatry, 12(3), 292-306. doi: 10.1038/sj.mp.4001943
Davalieva, K., Maleva Kostovska, I., & Dwork, A. J. (2016). Proteomics research in schizophrenia. Frontiers in Cellular Neuroscience, 10, 18. doi: 10.3389/fncel.2016.00018
Den Braber, A., Zilhão, N. R., Fedko, I. O., Hottenga, J. J., Pool, R., Smit, D. J. A., et al. (2016). Obsessive–compulsive symptoms in a large population-based twin-family sample are predicted by clinically based polygenic scores and by genome-wide SNPs. Translational Psychiatry, 6(2), 731. doi: 10.1038/tp.2015.223
Ehrnthaller, C., Ignatius, A., Gebhard, F., & Huber-Lang, M. (2011). New insights of an old defense system: Structure, function, and clinical relevance of the complement system. Molecular Medicine, 17(3), 317-29. PMID: 21046060
Filiou, M. D., Turck, C. W., & Martins-de-Souza, D. (2010). Quantitative proteomics for investigating psychiatric disorders. PROTEOMICS - Clinical Applications, 5(1-2), 38–49. doi: 10.1002/prca.201000060
Hasanzadeh, H., Rezaie-Tavirani, M., Seyyedi, S., & Emadi, A. (2015). [Proteomics study of extremely low frequency electromagnetic field (50 Hz) on human neuroblastoma cells (Persian)]. Koomesh, 17(1), 233-239.
Huang, T. L. (2002). Decreased serum albumin levels in Taiwanese patients with schizophrenia. Psychiatry and Clinical Neurosciences, 56(6), 627–30. doi: 10.1046/j.1440-1819.2002.01066.x
Jaffe, A. E., Deep-Soboslay, A., Tao, R., Hauptman, D. T., Kaye, W. H., Arango, V., et al. (2014). Genetic neuropathology of obsessive psychiatric syndromes. Translational Psychiatry, 4(9), 432. doi: 10.1038/tp.2014.68
Levin, Y., Wang, L., Schwarz, E., Koethe, D., Leweke, F. M., & Bahn, S. (2009). Global proteomic profiling reveals altered proteomic signature in schizophrenia serum. Molecular Psychiatry, 15(11), 1088–100. doi: 10.1038/mp.2009.54
Liu, S., Cui, J., Wang, X., Ma, J., Niu, H., Ma, X., et al. (2015). An association study between DLGAP1 rs11081062 and EFNA5 rs26728 polymorphisms with obsessive–compulsive disorder in a Chinese Han population. Neuropsychiatric Disease and Treatment, 11, 897-905. doi: 10.2147/ndt.s75009
Lin, Q., Cao, Y., & Gao, J. (2015).Decreased expression of the APOA1-APOC3-APOA4 gene cluster is associated with risk of Alzheimer's disease. Drug Design, Development and Therapy, 9, 5421-31. doi: 10.2147/dddt.s89279
Madhura, T. K. (2015). Role of serum cholesterol level in pathogenesis of OCD. Biochemistry & Analytical Biochemistry, 4(3), 1000183. doi: 10.4172/2161-1009.1000183
Mayilyan, K. R., Weinberger, D. R., & Sim, R. B. (2008). The complement system in schizophrenia. Drug News & Perspectives, 21(4), 200-10. doi: 10.1358/dnp.2008.21.4.1213349
Nejadi, N., Hantoushzadeh, S., Masti Mohammadpoor, S., Hosseini, Z. S., Mousavi, M., & Gorgani-Firuzjaee, S. (2015). Vitamin D Binding Protein as screening biomarker candidate for late-onset preeclampsia without intrauterine growth restriction during 16 week of gestation. Journal of Paramedical Sciences, 6(1), 36-43.
Noshirvani, H. F., Kasvikis, Y., Marks, I. M., Tsakiris, F., & Monteiro, W. O. (1991). Gender-divergent aetiological factors in obsessive-compulsive disorder. The British Journal of Psychiatry, 158(2), 260–263. doi: 10.1192/bjp.158.2.260
Pastore, N., Ballabio, A., & Brunetti-Pierri, N. (2013). Autophagy master regulator TFEB induces clearance of toxic SERPINA1/α-1-antitrypsin polymers. Autophagy, 9(7), 1094–6. doi: 10.4161/auto.24469
Qin, X., Swertfeger, D. K., Zheng, S., Hui, D. Y., & Tso, P. (1998). Apolipoprotein AIV: A potent endogenous inhibitor of lipid oxidation. American Journal of Physiology-Heart and Circulatory Physiology, 274(5), 1836-40. PMID: 9612397
Zamanian-Azodi, M., Rezaei-Tavirani, M., Rahmati-Rad, S., & Rezaei Tavirani, M. (2016). Ethanol and cancer induce similar changes on protein expression pattern of human fibroblast cell. Iranian Journal of Pharmaceutical Research, 15, 175-84. PMID: 28228815
Rezaei-Tavirani, M., Zamanian-Azodi, M., Rajabi, S., Masoudi-Nejad, A., Rostami-Nejad, M., & Rahmatirad, S. (2016). Protein Clustering and Interactome Analysis in Parkinson and Alzheimer’s Diseases. Archives of Iranian Medicine, 19(2), 101-9. doi: 0161902/AIM.007
Safaei, A., Rezaei Tavirani, M., Arefi Oskouei, A., Zamanian Azodi, M., Mohebbi, S. R., & Nikzamir, A. R. (2016). Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterology and Hepatology From Bed to Bench, 9(2), 114-23. PMID: 27099671
Samavat, S., Kalantari, S., Nafar, M., Rutishauser, D., Rezaei-Tavirani, M., Parvin, M., et al. (2015). Diagnostic urinary proteome profile for immunoglobulin a nephropathy. Iranian Journal of Kidney Diseases, 9(3), 239-48. PMID: 25957429
Şimşek, Ş., Gençoğlan, S., & Yüksel, T. (2016). DNA damage and antioxidants in treatment naïve children with obsessive–compulsive disorder. Psychiatry Research, 237, 133–7. doi: 10.1016/j.psychres.2016.01.054
Stewart, S. E., Jenike, M. A., & Keuthen, N. J. (2005). Severe obsessive-compulsive disorder with and without comorbid hair pulling. Journal of Clinical Psychiatry, 66(7), 864–9. doi: 10.4088/jcp.v66n0709
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth A., Minguez, P., et al. (2010). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research, 39, 561-8. PMID: 21045058
Tang, H. Y., Beer, L. A., & Speicher, D. W. (2011). In-Depth Analysis of a Plasma or Serum Proteome Using a 4D Protein Profiling Method. Serum/Plasma Proteomics, 728, 47–67. doi: 10.1007/978-1-61779-068-3_3
Yang, Y., Wan, C., Li, H., Zhu, H., La, Y., Xi, Z., et al. (2006). Altered levels of acute phase proteins in the plasma of patients with schizophrenia. Analytical Chemistry, 78(11), 3571–6. doi: 10.1021/ac051916x
Zali, H., Zamanian-Azodi, M., Rezaei Tavirani, M., & Akbar-Zadeh Baghban, A. (2015). Protein drug targets of lavandula angustifolia on treatment of rat Alzheimer’s disease. Iranian Journal of Pharmaceutical Research, 14(1), 291-302. PMID: 25561935
Zamanian-Azodi, M., Rezaei-Tavirani, M., Kermani-Ranjbar, T., Oskouie, A. A, RahmatiRad, S., & Asl, N. S. (2015). [Pathophysiology, genetics, types, and treatments in obsessive compulsive disorder (Persian)]. Koomesh, 16(4), 475-87.
Zilhão, N. R., Smit, D. J. A., den Braber, A., Dolan, C. V., Willemsen, G., Boomsma, D. I., & Cath, D. C. (2014). Genetic and environmental contributions to stability in adult obsessive compulsive behavior. Twin Research and Human Genetics, 18(1), 52–60. doi: 10.1017/thg.2014.77
Type of Study: Original |
Subject:
Behavioral Neuroscience
Received: 2016/11/16 | Accepted: 2017/04/12 | Published: 2017/07/1
Received: 2016/11/16 | Accepted: 2017/04/12 | Published: 2017/07/1
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |