Volume 10, Issue 4 (July & August 2019)
BCN 2019, 10(4): 401-408 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Rezaei Tavirani M, Rezaei Tavirani S, Zadeh-Esmaeel M, Ahmadi N A. Introducing Critical Pain-related Genes: A System Biology Approach. BCN 2019; 10 (4) :401-408
URL: http://bcn.iums.ac.ir/article-1-1328-en.html
URL: http://bcn.iums.ac.ir/article-1-1328-en.html
Mostafa Rezaei Tavirani * 
 1, Sina Rezaei Tavirani1
1, Sina Rezaei Tavirani1 
 , Mohammad-Mahdi Zadeh-Esmaeel1
, Mohammad-Mahdi Zadeh-Esmaeel1 
 , Nayeb Ali Ahmadi1
, Nayeb Ali Ahmadi1 


 1, Sina Rezaei Tavirani1
1, Sina Rezaei Tavirani1 
 , Mohammad-Mahdi Zadeh-Esmaeel1
, Mohammad-Mahdi Zadeh-Esmaeel1 
 , Nayeb Ali Ahmadi1
, Nayeb Ali Ahmadi1 

1- Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Full-Text [PDF 3579 kb]
| Abstract (HTML)
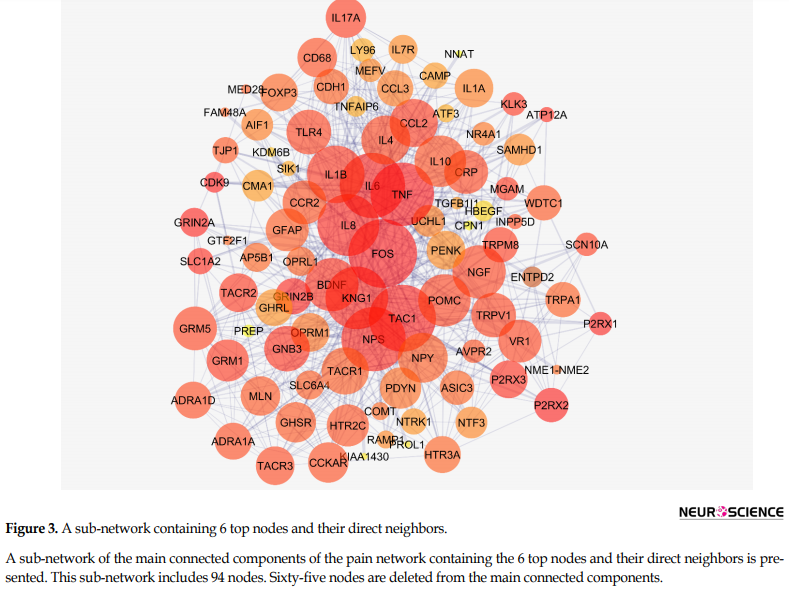
Figure 2 shows the connections between the six top nodes of the pain network in the represented sub-network. Considering the dense relationship between these central nodes, it seems these 6 genes and their direct neighbors play a critical role in the integrity of the pain network. A sub-network, including the 6 central nodes and their direct neighbors is constructed and is shown in Figure 3. Since the six central nodes have direct access to the 88 nodes, we examined the least number of central nodes which are necessary to maintain the integrity of the sub-network. After using different combinations of the six genes, it was found that FOS, IL6, TAC1, and KNG1 combination is the best grouping which serves as the core of the sub-network (Figure 4).
Emma V. Bird et al. investigation about the role of Chemokine Lymphotactin (XCL1) and its receptor (XCR1) in the nervous system led to introduce the role of FOS in pain. They reported that expression of c-FOS in trigeminal subnucleus caudalis exposed to XCL1 increased. Based on their analysis, XCL1-XCR1 axis is involved in the peripheral and central trigeminal pain pathway. In other words, induction of c-FOS, extracellular signal-regulated kinase, and PP38 expression and also hyper-excitability within the trigeminal subnucleus caudalis nociceptive circuitry compose the possible proposed mechanism (Bird et al., 2018).
The second central gene is interleukin 6. It is a well-known cytokine which plays different roles in host defense because of its vast range of activities in immune and hematopoietic processes. It is considered as one of the key elements in acute phase response induction. Many diseases are associated with over-expression of IL-6 (Simpson, Hammacher, Smith, Matthews, & Ward, 1997). Zhou et al. (2016) published a review of the regulatory role of IL6 in pathological pain. Based on numerous documents, they stated the essential part for IL6 in pathological types of pain. They investigated the importance of IL6 in the pathological types of pain associated with several diseases such as bone cancer, the peripheral neuropathy due to chemotherapy, and spinal cord and peripheral nerve injuries (Zhou et al., 2016).
Tachykinins encode small polypeptide chains which are known as different neuropeptides. TAC1, TAC3, and TAC4 belong to this gene family. It was established that TAC1 contributes to inflammation and immune response (Botz et al., 2013). Reportedly, TAC1 encodes neuropeptide Substance P (SP) which is involved in the neurogenic inflammation process. Up-regulation of SP in A and B neurons after noxious stimulation and its presence in C fibers indicate the role of TAC1 gene product in inflammatory pain (Shanley, Lear, Davidson, Ross, MacKenzie, 2011).
There is evidence that bradykinin is involved in sensitizing nervous (nociceptor) peripheral terminals, which lead to a reduction of pain threshold (Wang et al., 2005). The investigation is shown that there are two homologous copies; KNG1 and KNG2 as kininogen gene which High-molecular-weight Kininogen (HK) is a product of KIN1-derived mRNA. Bradykinin results from cleaved HK (Yang et al., 2017). Absence of pain due to SCN9A mutation and also a spectrum of human genetic pain disorders were reported (Drenth & Waxman, 2007; Rajasekharan, Martens, Domingues, & Cauwels, 2017).It seems that the introduced critical genes alone or in a combination are suitable candidates for drug targeting.
Network analysis showed six critical genes which are involved in pain development. Considering up-regulation of the key genes, suitable inhibitor reagents of the introduced important genes can be attentive as painkillers.
Ethical Considerations
Compliance with ethical guidelines
This research is confirmed by ethical Code of IR.SBMU.RETECH.REC.1397.318 by Shahid Beheshti University of Medical Sciences.
Funding
Shahid Beheshti University of Medical Sciences financially supported the research.
Authors' contributions
Involved in the conceptualization of the study and original draft preparation of the manuscript: Mostafa Rezaei-Tavirani, Nayeb Ali Ahmadi and Mohammad-Mahdi Zadeh-Esmaeel; Carried out the technical studies and data analysis: Sina Rezaei-Tavirani and Mostafa Rezaei-Tavirani; Participated in the writing the final version of the manuscript: all authors.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgments
We acknowledge the support of Shahid Beheshti University of Medical Sciences.
References
Full-Text:
1. Introduction
espite many investigations on the types of pain, unfortunately, the concept of pain is still unclear. This problem has resulted from the absence of distinction between pain sensation and its causes. Pain is a sensation, and this sense has several common features with the other sensations such as itching. It also occurs in various locations of the body. Due to this complexity, types of pain are categorized based on location, etiology, intensity, duration, and pathophysiology. However, pain cannot be considered just a physical entity but a sensation as well. Types of pain are known as distressing experiences, which are associated with damage or injury to the tissues.
Pain has various dimensions, such as sensory, emotional, cognitive, and social (Brodal, 2017; Orr, Shank, & Black, 2017; Williams & Craig, 2016). Based on the evidence, genetics has an important impact on pain sensation. So far, several genes have been introduced that affect pain sensation and its intensity. Reportedly, people respond differently to painful stimuli. Genetics may provide reasons for such different reactions between patients. Investigation shows that pain development is predictable. It seems that better understanding of the molecular mechanism of pain can mark suitable drug targets and more potent pain killers (Fillingim, Wallace, Herbstman, Ribeiro‐Dasilva, & Staud, 2008; Foulkes & Wood, 2008). This genetic variability in individuals calls for continuous efforts to achieve effective treatment of pain. Genome-wide investigation and bioinformatics can be used to gain a new perspective about types of pain (Clarke et al., 2015; Smith et al., 2018).
Several therapeutic guidelines for types of pain based on reported phenotypes have been established. Some efforts are made to collect dispersed documents to present a gene set responsible for variation in pain sensation (Van Hecke et al., 2015; Veluchamy, Hébert, Meng, Palmer, & Smith, 2018). Protein-Protein Interaction (PPI) network has drawn the attention of scientists to solve such genetic complexity, which hopefully explains the types of pain. In this approach, the all known genes related to specific diseases are included in an interacting unit while each plays different roles relative to the others in the integrity of the constructed network. The important genes are highlighted as central genes and can be considered as prominent genes in the onset or development of the disorder (Safari-Alighiarloo, Taghizadeh, Rezaei-Tavirani, Goliaei, & Peyvandi, 2014). In the present study, a network analysis approach for pain is planned to introduce critically involved genes in different types of pain.
2. Methods
STRING database (Szklarczyk et al., 2016) was used as genes resource. The genes related to pain were extracted via disease query of the database. The genes were included in a PPI network by using Cytoscape software version 3.6.0 (Tavirani et al., 2018). Network analyzer a plugin of Cytoscape software was applied to determine the central nodes of the network. The top nodes (the nodes with closeness above mean +2 SD) were identified (Safari‐Alighiarloo et al., 2017). A sub-network, including the central nodes and their direct neighbors, was constructed, too. A combination containing the least number of central nodes that were necessary to maintain the integrity of this sub-network was identified. To consider the other nodes which were not included in the neighbor nodes sub-network, they interacted in the non-neighbors nodes sub-network, and the critical nodes based on degree value were identified. The central nodes of both sub-networks were considered as key genes related to pain.
3. Results
A total number of 214 genes related to types of pain were extracted and included in the interactome. The network of 49 isolated genes, 2 triple units, and a main connected component (characterized by 159 nodes) was constructed (Figure 1). Closeness is defined as 1/avg L(n, m), where avg stands for average and L(n, m) is the length of the shortest path between two nodes n and m, so it is a measure of how fast information spreads from a given node to other reachable nodes in network (Malhotra, Jha, Singh, & Pandey, 2017). Therefore the nodes with the higher value of degree (or higher amount of neighbors) can be considered as the high score nodes based on closeness value. The six top nodes (FOS, IL6, TNF, TAC1, IL8, and KNG1) based on closeness value considering the cut-off value of Mean±SD were determined and presented in Table 1. In this Figure, degree, length of the shortest path between two nodes n and m, betweenness centrality, and closeness centrality related to the six central nodes are shown.
espite many investigations on the types of pain, unfortunately, the concept of pain is still unclear. This problem has resulted from the absence of distinction between pain sensation and its causes. Pain is a sensation, and this sense has several common features with the other sensations such as itching. It also occurs in various locations of the body. Due to this complexity, types of pain are categorized based on location, etiology, intensity, duration, and pathophysiology. However, pain cannot be considered just a physical entity but a sensation as well. Types of pain are known as distressing experiences, which are associated with damage or injury to the tissues.
Pain has various dimensions, such as sensory, emotional, cognitive, and social (Brodal, 2017; Orr, Shank, & Black, 2017; Williams & Craig, 2016). Based on the evidence, genetics has an important impact on pain sensation. So far, several genes have been introduced that affect pain sensation and its intensity. Reportedly, people respond differently to painful stimuli. Genetics may provide reasons for such different reactions between patients. Investigation shows that pain development is predictable. It seems that better understanding of the molecular mechanism of pain can mark suitable drug targets and more potent pain killers (Fillingim, Wallace, Herbstman, Ribeiro‐Dasilva, & Staud, 2008; Foulkes & Wood, 2008). This genetic variability in individuals calls for continuous efforts to achieve effective treatment of pain. Genome-wide investigation and bioinformatics can be used to gain a new perspective about types of pain (Clarke et al., 2015; Smith et al., 2018).
Several therapeutic guidelines for types of pain based on reported phenotypes have been established. Some efforts are made to collect dispersed documents to present a gene set responsible for variation in pain sensation (Van Hecke et al., 2015; Veluchamy, Hébert, Meng, Palmer, & Smith, 2018). Protein-Protein Interaction (PPI) network has drawn the attention of scientists to solve such genetic complexity, which hopefully explains the types of pain. In this approach, the all known genes related to specific diseases are included in an interacting unit while each plays different roles relative to the others in the integrity of the constructed network. The important genes are highlighted as central genes and can be considered as prominent genes in the onset or development of the disorder (Safari-Alighiarloo, Taghizadeh, Rezaei-Tavirani, Goliaei, & Peyvandi, 2014). In the present study, a network analysis approach for pain is planned to introduce critically involved genes in different types of pain.
2. Methods
STRING database (Szklarczyk et al., 2016) was used as genes resource. The genes related to pain were extracted via disease query of the database. The genes were included in a PPI network by using Cytoscape software version 3.6.0 (Tavirani et al., 2018). Network analyzer a plugin of Cytoscape software was applied to determine the central nodes of the network. The top nodes (the nodes with closeness above mean +2 SD) were identified (Safari‐Alighiarloo et al., 2017). A sub-network, including the central nodes and their direct neighbors, was constructed, too. A combination containing the least number of central nodes that were necessary to maintain the integrity of this sub-network was identified. To consider the other nodes which were not included in the neighbor nodes sub-network, they interacted in the non-neighbors nodes sub-network, and the critical nodes based on degree value were identified. The central nodes of both sub-networks were considered as key genes related to pain.
3. Results
A total number of 214 genes related to types of pain were extracted and included in the interactome. The network of 49 isolated genes, 2 triple units, and a main connected component (characterized by 159 nodes) was constructed (Figure 1). Closeness is defined as 1/avg L(n, m), where avg stands for average and L(n, m) is the length of the shortest path between two nodes n and m, so it is a measure of how fast information spreads from a given node to other reachable nodes in network (Malhotra, Jha, Singh, & Pandey, 2017). Therefore the nodes with the higher value of degree (or higher amount of neighbors) can be considered as the high score nodes based on closeness value. The six top nodes (FOS, IL6, TNF, TAC1, IL8, and KNG1) based on closeness value considering the cut-off value of Mean±SD were determined and presented in Table 1. In this Figure, degree, length of the shortest path between two nodes n and m, betweenness centrality, and closeness centrality related to the six central nodes are shown.
Figure 2 shows the connections between the six top nodes of the pain network in the represented sub-network. Considering the dense relationship between these central nodes, it seems these 6 genes and their direct neighbors play a critical role in the integrity of the pain network. A sub-network, including the 6 central nodes and their direct neighbors is constructed and is shown in Figure 3. Since the six central nodes have direct access to the 88 nodes, we examined the least number of central nodes which are necessary to maintain the integrity of the sub-network. After using different combinations of the six genes, it was found that FOS, IL6, TAC1, and KNG1 combination is the best grouping which serves as the core of the sub-network (Figure 4).
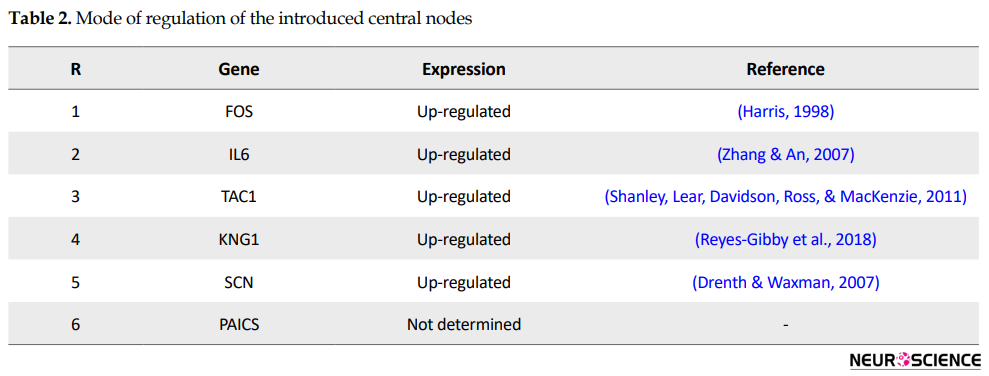
This sub-network includes 88 nodes (2 excluded central node+86 direct neighbors). In this procedure, the number of nodes which were not included in Figure 3 was not considered. However, these vanished genes may play roles in types of pain. So the missing genes were included in a sub-network as it is shown in Figure 5. Sub-network analysis revealed that SNC8A, SNC9A, SNC11A, (SNCA gene family) and PAICS are the hubs based on the cut-off value of Mean±SD. Mode of regulation of the 4 central nodes and SNCA gene family was investigated and shown in Table 2.
4. Discussion
Based on the evidence, the genes related to specific diseases need additional relevant genes to construct a scale-free PPI network. Network analysis can screen the interacted genes to find important ones which may be useful in disease management (Safari-Alighiarloo et al., 2014). As it is shown in Figure 1 and Table 1, six central nodes are introduced, which can be considered as the key elements in pain. The findings that are presented in the figures 2, 3 , and 4 refer to the significant role of FOS, IL6, TAC1, and KNG1 among the six central genes in control of pain PPI network.
Further analysis (Figure 5) also revealed the noteworthy role of SNC8, SNC9, SNC11A, and PAICS in pain. It is well-known that SNC8A, SNC9A, and SNC11A are varieties of voltage-gated channel alpha subunit (SNCA) (England & De Groot, 2009). It seems that FOS, IL6, TAC1, KNG1, SNCA, and PAICS are the crucial genes related to the types of pain. As it is shown in Table 2, all crucial genes are up-regulated (a direct statement about the regulation of PAICS was not found in the literature). The possible roles of the introduced critical genes in pain are discussed briefly in the following paragraphs.
Based on the evidence, the genes related to specific diseases need additional relevant genes to construct a scale-free PPI network. Network analysis can screen the interacted genes to find important ones which may be useful in disease management (Safari-Alighiarloo et al., 2014). As it is shown in Figure 1 and Table 1, six central nodes are introduced, which can be considered as the key elements in pain. The findings that are presented in the figures 2, 3 , and 4 refer to the significant role of FOS, IL6, TAC1, and KNG1 among the six central genes in control of pain PPI network.
Further analysis (Figure 5) also revealed the noteworthy role of SNC8, SNC9, SNC11A, and PAICS in pain. It is well-known that SNC8A, SNC9A, and SNC11A are varieties of voltage-gated channel alpha subunit (SNCA) (England & De Groot, 2009). It seems that FOS, IL6, TAC1, KNG1, SNCA, and PAICS are the crucial genes related to the types of pain. As it is shown in Table 2, all crucial genes are up-regulated (a direct statement about the regulation of PAICS was not found in the literature). The possible roles of the introduced critical genes in pain are discussed briefly in the following paragraphs.
Emma V. Bird et al. investigation about the role of Chemokine Lymphotactin (XCL1) and its receptor (XCR1) in the nervous system led to introduce the role of FOS in pain. They reported that expression of c-FOS in trigeminal subnucleus caudalis exposed to XCL1 increased. Based on their analysis, XCL1-XCR1 axis is involved in the peripheral and central trigeminal pain pathway. In other words, induction of c-FOS, extracellular signal-regulated kinase, and PP38 expression and also hyper-excitability within the trigeminal subnucleus caudalis nociceptive circuitry compose the possible proposed mechanism (Bird et al., 2018).
The second central gene is interleukin 6. It is a well-known cytokine which plays different roles in host defense because of its vast range of activities in immune and hematopoietic processes. It is considered as one of the key elements in acute phase response induction. Many diseases are associated with over-expression of IL-6 (Simpson, Hammacher, Smith, Matthews, & Ward, 1997). Zhou et al. (2016) published a review of the regulatory role of IL6 in pathological pain. Based on numerous documents, they stated the essential part for IL6 in pathological types of pain. They investigated the importance of IL6 in the pathological types of pain associated with several diseases such as bone cancer, the peripheral neuropathy due to chemotherapy, and spinal cord and peripheral nerve injuries (Zhou et al., 2016).
Tachykinins encode small polypeptide chains which are known as different neuropeptides. TAC1, TAC3, and TAC4 belong to this gene family. It was established that TAC1 contributes to inflammation and immune response (Botz et al., 2013). Reportedly, TAC1 encodes neuropeptide Substance P (SP) which is involved in the neurogenic inflammation process. Up-regulation of SP in A and B neurons after noxious stimulation and its presence in C fibers indicate the role of TAC1 gene product in inflammatory pain (Shanley, Lear, Davidson, Ross, MacKenzie, 2011).
There is evidence that bradykinin is involved in sensitizing nervous (nociceptor) peripheral terminals, which lead to a reduction of pain threshold (Wang et al., 2005). The investigation is shown that there are two homologous copies; KNG1 and KNG2 as kininogen gene which High-molecular-weight Kininogen (HK) is a product of KIN1-derived mRNA. Bradykinin results from cleaved HK (Yang et al., 2017). Absence of pain due to SCN9A mutation and also a spectrum of human genetic pain disorders were reported (Drenth & Waxman, 2007; Rajasekharan, Martens, Domingues, & Cauwels, 2017).It seems that the introduced critical genes alone or in a combination are suitable candidates for drug targeting.
Network analysis showed six critical genes which are involved in pain development. Considering up-regulation of the key genes, suitable inhibitor reagents of the introduced important genes can be attentive as painkillers.
Ethical Considerations
Compliance with ethical guidelines
This research is confirmed by ethical Code of IR.SBMU.RETECH.REC.1397.318 by Shahid Beheshti University of Medical Sciences.
Funding
Shahid Beheshti University of Medical Sciences financially supported the research.
Authors' contributions
Involved in the conceptualization of the study and original draft preparation of the manuscript: Mostafa Rezaei-Tavirani, Nayeb Ali Ahmadi and Mohammad-Mahdi Zadeh-Esmaeel; Carried out the technical studies and data analysis: Sina Rezaei-Tavirani and Mostafa Rezaei-Tavirani; Participated in the writing the final version of the manuscript: all authors.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgments
We acknowledge the support of Shahid Beheshti University of Medical Sciences.
References
- Bird, E. V., Iannitti, T., Christmas, C. R., Obara, I., Andreev, V. I., King, A. E., et al. (2018). A novel role for Lymphotactin (XCL1) signaling in the nervous system: XCL1 acts via its receptor XCR1 to Increase trigeminal neuronal excitability. Neuroscience, 379, 334-49. [DOI:10.1016/j.neuroscience.2018.03.030] [PMID] [PMCID]
- Botz, B., Imreh, A., Sándor, K., Elekes, K., Szolcsányi, J., Reglődi, D., et al. (2013). Role of pituitary adenylate-cyclase activating polypeptide and Tac1 gene derived tachykinins in sensory, motor and vascular functions under normal and neuropathic conditions. Peptides, 43, 105-12. [DOI:10.1016/j.peptides.2013.03.003] [PMID]
- Brodal, P. (2017). A neurobiologist’s attempt to understand persistent pain. Scandinavian Journal of Pain, 15(1), 140-7. [DOI:10.1016/j.sjpain.2017.03.001] [PMID]
- Clarke, H., Katz, J., Flor, H., Rietschel, M., Diehl, S. R., & Seltzer, Z. E. (2015). Genetics of chronic post-surgical pain: A crucial step toward personal pain medicine. Canadian Journal of Anesthesia, 62(3), 294-303. [DOI:10.1007/s12630-014-0287-6] [PMID]
- Drenth, J. P., & Waxman, S. G. (2007). Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. The Journal of Clinical Investigation, 117(12), 3603-9. [DOI:10.1172/jci33297] [PMID] [PMCID]
- England, S., & De Groot, M. J. (2009). Subtype‐selective targeting of voltage‐gated sodium channels. British Journal of Pharmacology, 158(6), 1413-25. [DOI:10.1111/j.1476-5381.2009.00437.x] [PMID] [PMCID]
- Fillingim, R. B., Wallace, M. R., Herbstman, D. M., Ribeiro‐Dasilva, M., & Staud, R. (2008). Genetic contributions to pain: A review of findings in humans. Oral Diseases, 14(8), 673-82. [DOI:10.1111/j.1601-0825.2008.01458.x] [PMID] [PMCID]
- Foulkes, T., & Wood, J. N. (2008). Pain genes. PLOS Genetics, 4(7), e1000086. [DOI:10.1371/journal.pgen.1000086] [PMID] [PMCID]
- Harris, J. A. (1998). Using c-fos as a neural marker of pain. Brain Research Bulletin, 45(1), 1-8. [DOI:10.1016/s0361-9230(97)00277-3]
- Malhotra, A. G., Jha, M., Singh, S., & Pandey, K. M. (2017). Construction of a comprehensive protein–protein interaction map for vitiligo disease to identify key regulatory elements: A systemic approach. Interdisciplinary Sciences, 10(3):500-14. [DOI:10.1007/s12539-017-0213-z] [PMID]
- Orr, P. M., Shank, B. C., & Black, A. C. (2017). The role of pain classification systems in pain management. Critical Care Nursing Clinics, 29(4), 407-18. [DOI:10.1016/j.cnc.2017.08.002] [PMID]
- Rajasekharan, S., Martens, L., Domingues, L., & Cauwels, R. (2017). SCN9A channelopathy associated autosomal recessive Congenital Indifference to Pain. European Journal of Paediatric Dentistry, 18(1), 66-8. [DOI:10.23804/ejpd.2017.18.01.14] [PMID]
- Reyes-Gibby, C. C., Wang, J., Yeung, S. C. J., Chaftari, P., Robert, K. Y., Hanna, E. Y., et al. (2018). Genome-wide association study identifies genes associated with neuropathy in patients with head and neck cancer. Scientific Reports, 8(1), 8789. [DOI:10.1038/S41598-018-27070-4] [PMID] [PMCID]
- Safari-Alighiarloo, N., Taghizadeh, M., Rezaei-Tavirani, M., Goliaei, B., & Peyvandi, A. A. (2014). Protein-Protein Interaction networks (PPI) and complex diseases. Gastroenterology and Hepatology from Bed to Bench, 7(1), 17-31. [PMID] [PMCID]
- Safari‐Alighiarloo, N., Taghizadeh, M., Tabatabaei, S. M., Shahsavari, S., Namaki, S., Khodakarim, S., et al. (2017). identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes. Journal of Diabetes, 9(8), 764-77. [DOI:10.1111/1753-0407.12483] [PMID]
- Shanley, L., Lear, M., Davidson, S., Ross, R., & MacKenzie, A. (2011). Evidence for regulatory diversity and auto-regulation at the TAC1 locus in sensory neurones. Journal of Neuroinflammation, 8, 10. [DOI:10.1186/1742-2094-8-10] [PMID] [PMCID]
- Simpson, R. J., Hammacher, A., Smith, D. K., Matthews, J. M., & Ward, L. D. (1997). Interleukin‐6: Structure‐function relationships. Protein Science, 6(5), 929-55. [DOI:10.1002/PRO.5560060501] [PMID] [PMCID]
- Smith, S., Parisien, M., Gris, P., Khoury, S., Slade, G., Bair, E., et al. (2018). Genome-wide DNA methylation analysis and integrative bioinformatics in the OPPERA study of painful TMD. The Journal of Pain, 19(3), S100. [DOI:10.1016/J.JPAIN.2017.12.224]
- Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2016). The STRING database in 2017: Quality-controlled protein- protein association networks, made broadly accessible. Nucleic Acids Research, 45(D1), D362-D8. [DOI:10.1093/NAR/GKW937] [PMID] [PMCID]
- Tavirani, M. R., Bashash, D., Rostami, F. T., Tavirani, S. R., Nikzamir, A., Tavirani, M. R., et al. (2018). Celiac disease microarray analysis based on system biology approach. Gastroenterology and Hepatology from Bed to Bench, 11(3), 216-24. [PMID] [PMCID]
- Van Hecke, O., Kamerman, P. R., Attal, N., Baron, R., Bjornsdottir, G., Bennett, D. L., et al. (2015). Neuropathic Pain Phenotyping by International Consensus (NeuroPPIC) for genetic studies: A NeuPSIG systematic review, Delphi survey, and expert panel recommendations. Pain, 156(11), 2337-53. [DOI:10.1097/J.PAIN.0000000000000335] [PMID] [PMCID]
- Veluchamy, A., Hébert, H. L., Meng, W., Palmer, C. N., & Smith, B. H. (2018). Systematic review and meta-analysis of genetic risk factors for neuropathic pain. Pain, 159(5), 825-48. [DOI:10.1097/J.PAIN.0000000000001164] [PMID]
- Wang, H., Kohno, T., Amaya, F., Brenner, G. J., Ito, N., Allchorne, A., et al. (2005). Bradykinin produces pain hypersensitivity by potentiating spinal cord glutamatergic synaptic transmission. Journal of Neuroscience, 25(35), 7986-92. [DOI:10.1523/JNEUROSCI.2393-05.2005] [PMID]
- Williams, A. C., & Craig, K. D. (2016). Updating the definition of pain. Pain, 157(11), 2420-3. [DOI:10.1097/J.PAIN.0000000000000613] [PMID]
- Yang, A., Xie, Z., Wang, B., Colman, R. W., Dai, J., & Wu, Y. (2017). An essential role of high-molecular-weight kininogen in endotoxemia. Journal of Experimental Medicine, 214(9), 2649-70. [DOI:10.1084/JEM.20161900] [PMID] [PMCID]
- Zhang, J. M., & An, J. (2007). Cytokines, inflammation and pain. International Anesthesiology Clinics, 45(2), 27-37. [DOI:10.1097/aia.0b013e318034194e] [PMID] [PMCID]
- Zhou, Y. Q., Liu, Z., Liu, Z. H., Chen, S. P., Li, M., Shahveranov, A., et al. (2016). Interleukin-6: An emerging regulator of pathological pain. Journal of Neuroinflammation, 13, 141. [DOI:10.1186/s12974-016-0607-6] [PMID] [PMCID]
Type of Study: Original |
Subject:
Behavioral Neuroscience
Received: 2018/09/5 | Accepted: 2019/01/10 | Published: 2019/07/1
Received: 2018/09/5 | Accepted: 2019/01/10 | Published: 2019/07/1
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |