BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://bcn.iums.ac.ir/article-1-1106-en.html
2- Department of Applied Cell Sciences, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran.
3- Department of Biological Sciences, Faculty of Biological Sciences and Technologies, Shahid Beheshti University, Tehran, Iran.
1. Introduction
SHANK3 (SH3 and multiple ankyrin repeat domain 3) is one of the most promising genes involved in neuronal function. SHANK3 protein is a vital scaffolding protein of the Postsynaptic Density (PSD) of excitatory glutamatergic synapses in the Central Nervous System (CNS) (Kreienkamp, 2008). The gene is located on chromosome 22q13.3, spans about 58 kb, and comprises 24 exons, 7 of which are alternatively spliced (Durand et al., 2007). The product of SHANK3 gene expression is a large protein containing diverse domains such as an ANK repeat, a proline-rich cluster, SH3, PDZ, and SAM (sterile a-motif) (Kreienkamp, 2008). This molecule directly or indirectly interacts with cytoskeletal proteins and neurotransmitter receptors. It is also involved in the formation, expansion, and development of dendritic spines and is an essential factor for developing functional synapses (Roussignol et al., 2005). Different mutations in the sequence of the SHANK3 gene may disrupt the structure and function of gene domains (Mameza et al., 2013), and the resulting variants may be associated with various brain disorders. Non-synonymous/frame-shift and deletion/duplication mutations in SHANK3 have been detected in PD patients (Wilson et al., 2003). It has been reported that a non-synonymous polymorphism named rs9616915 (T>C) may occur in the exon 6 of the SHANK3 gene, which induces an amino acid replacement, i.e. isoleucine to threonine. This alteration directly affects the gene function of splicing regulation and results in protein structure damage (Shao et al., 2014). In this study, we aimed to evaluate the association between (rs9616915) SHANK3 polymorphism and susceptibility to Parkinson disease in people living in the north of Iran.
2. Materials and Methods
A total of 100 patients with PD (76 males) and 100 healthy controls participated in the study. The controls and patients were unrelated subjects selected from the same population living in Guilan Province, north of Iran. The patients’ characteristics were collected from Iran Medical Diagnostic Center in Rasht City, Iran. The subjects’ age ranges between 60 and 80 years. The participants were investigated in terms of having certain genetic diseases in close relatives. Also, they would be excluded from the study if they had a history of any other neurological disorder, acquired brain injury, or severe mental illness. Each subject gave a 2-mL blood sample drawn into Ethylenediaminetetraacetic Acid (EDTA)-coated tubes (Venoject, Belgium) for the next step of genomic DNA extraction. The local Ethics Committee has approved this study, and written consent was obtained from all participants.
2.2. Genotyping
Genomic DNA was extracted from whole-blood samples using DNA Extractor Gpp Solution Kit (Gen Pajoohan, Iran) according to the manufacturer’s instructions. The intended SNP (rs9616915 in the SHANK3 gene) was evaluated by polymerase chain reaction-restriction fragment length polymorphism assay. Primer sequences for PCR reactions were 5’-GCTTGACACCCCTCTACCA -3’ (forward) and 5’ -TCTGCCCCATAGAACAGC-3’ (reverse). PCR reactions were performed in a total volume of 25 µL and consisted of an initial denaturation step at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 30 s and annealing at 55°C for 30 s. PCR products were subsequently digested by restriction enzyme hphl. After endonuclease digestion, products were separated on 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
2.3. Statistical analysis
Statistical analysis was performed by MedCalc (version12.1, Mariakerke, Belgium). Analysis of differences in allele and genotype frequencies between cases and controls was done using the χ2 test. To estimate the association between the SHANK3 rs9616915 variant and the risk of Parkinson disease, the odds ratios with 95% confidence intervals (95% CI) were evaluated by logistic regression. A value of P<0.05 was considered statistically significant.
3. Results
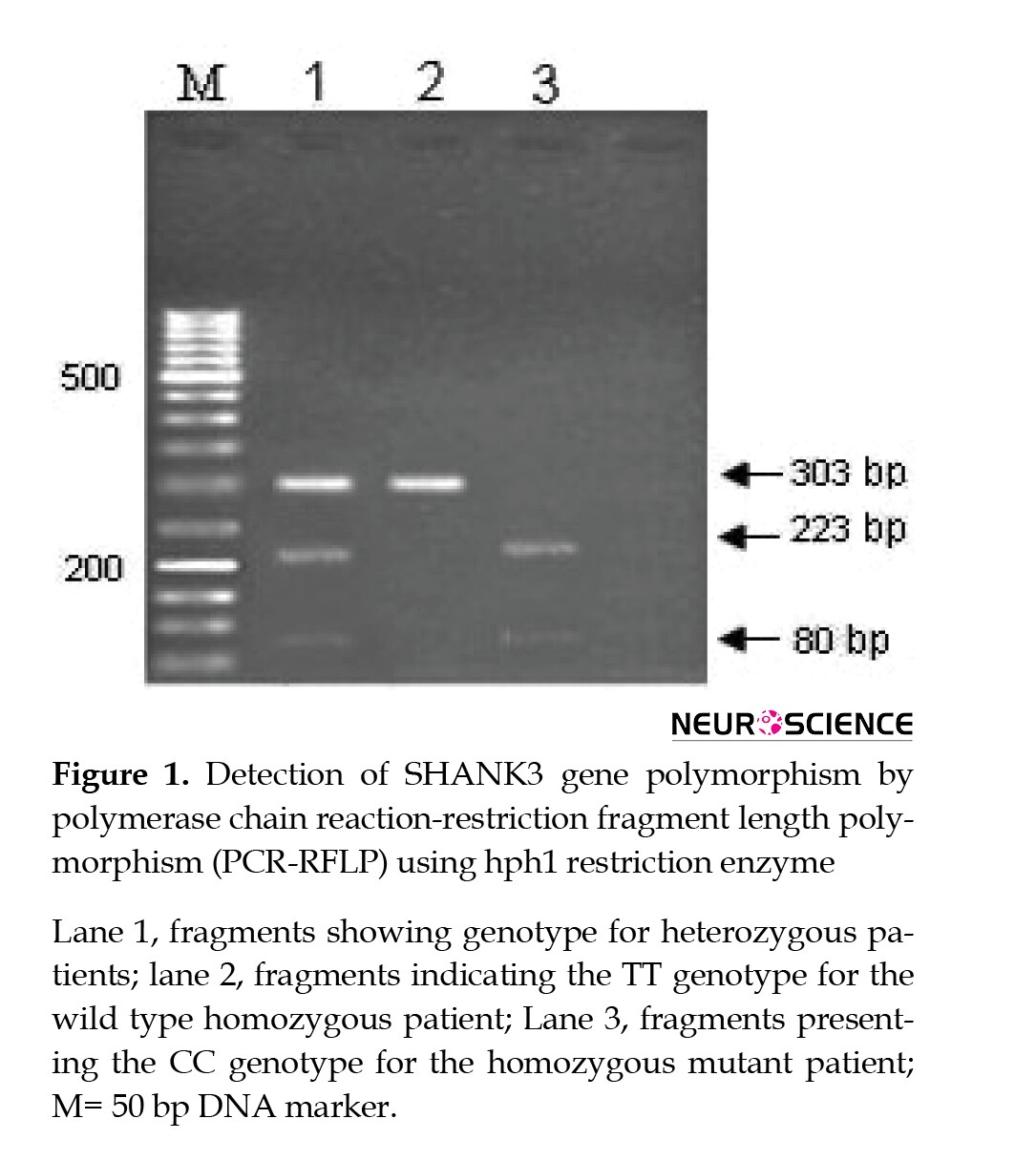
Genotype frequencies for CC, TC, and TT were 33%, 50%, and 17% in controls, and 24%, 72%, and 4% in PD patients, respectively. Statistical analysis showed a significant difference between the two groups (P=0.001). The results indicated that the subgroup with TC genotype had an increased risk of Parkinson disease (P=0.035, OR=1.98, 95% CI=1.04 - 3.74). All information about allele and genotype frequencies and associated ORs (95% CI) for cases and controls are summarized in Table 1.
.jpg)
Besides, as most of our cases were male subjects (76 out of 100), we re-analyzed the data again for male cases. After removing the female genotypes from our population, the statistical analysis revealed that the results are consistent with females. The individuals with TC genotype had an increased risk of Parkinson disease (P=0.029, OR=2.17, 95% CI=1.08 – 4.37) (Table 2).
.jpg)
4. Discussion
SHANK3 is a member of the highly conserved ProSAP/Shank family of master synaptic scaffolding proteins (Qualmann, Boeckers, Jeromin, Gundelfinger, & Kessels, 2004; Bozdagi et al., 2010). The reduced expression of SHANK3 results in the attenuation of the number of dendrites and synaptic transmission and plasticity. Malfunctions of synaptic transmission might result in brain disorders such as epilepsy, autism, Alzheimer, and Parkinson disease (Lepeta et al., 2016). It has been shown that the loss of SHANK3 in in vitro cultures of hippocampal neurons may decrease the number and augmented length of dendritic spines. In contrast, expression of SHANK3 could induce the formation of de novo functional dendritic spines in aspiny cerebellar neurons (Roussignol et al., 2005).
Moreover, the absence of one copy of the SHANK3 gene or its mutations could result in changes in spine morphology and behavioral issues related to autism traits such as deficits in social communication, social interaction, and repetitive actions (Bangash et al., 2011; Durand et al., 2012). Accomplished studies about this gene have indicated that genetic variations of SHANK3 play an essential role in susceptibility to many kinds of neurodevelopmental disorders. Meanwhile, rs9616915 polymorphism is a non-synonymous substitution from isoleucine to threonine in the exon 6 of SHANK3, which could interfere with the gene function via affecting the splicing process (Shao et al., 2014). Previous studies have reported its functional involvement in hippocampal mRNA expression of SHANK3 (Zhang, Wu, Hong, Peng, & Fang, 2016). In a case-control study in China, it was reported that rs9616915 polymorphism is associated with decreased risk of Autism Spectrum Disorders (ASD) (Shao et al., 2014). However, other studies have not found an association between rs9616915 and ASD (Qin et al., 2009; Sykes et al., 2009; Jonsson et al., 2014). In another study, Mashayekhi, Mizban, Bidabadi, and Salehi (2016) reported that rs9616915 is accompanied by a higher risk of ASD.
The inconsistency may be related to different environmental conditions, relatively small sample size, variations in diagnostic criteria of the disorder, and diverse ethnicity. For example, it has been stated that the C allele frequency is 48.1% in the European population while it is 3.5% among Han Chinese people (Zhang et al., 2016). In other brain disorders, it has been shown that the synaptic dysfunction and loss of glutamate receptors at the Shank-postsynaptic platform could contribute to Alzheimer disease (Gong, Lippa, Zhu, Lin, & Rosso, 2009). A significant association between SHANK3 variation and Phelan-McDermid syndrome has also been reported (Bonaglia et al., 2011). Moreover, mutations in the SHANK3 gene are associated with schizophrenia (Gauthier et al., 2010). This mutation in bipolar disorder has also been investigated in previous studies (Zhang et al., 2016). These findings led us to hypothesize that a functional polymorphism within the SHANK3 gene might be linked to susceptibility to Parkinson disease. To our knowledge, this is the first study evaluating the association between SHANK3 gene polymorphism and the risk of Parkinson disease. However, it should be mentioned that this study has some limitations. We have only evaluated one SNP in the SHANK3 gene, which is not adequate to assess the risk of Parkinson disease in a subject, and many other genes and SNPs may also be involved in the risk of disease. Besides, our sample size was not so large, and further studies with larger sample sizes are needed in the future to confirm the role of the SHANK3 gene in Parkinson disease.
5. Conclusion
Ethical Considerations
Compliance with ethical guidelines
Funding
Authors' contributions
Conflict of interest
Reference
Bangash, M. A., Park, J. M., Melnikova, T., Wang, D., Jeon, S. K., & Lee, D., et al. (2011). RETRACTED: Enhanced polyubiquitination of shank3 and NMDA receptor in a mouse model of Autism. Cell, 145(5), 758-72. [DOI:10.1016/j.cell.2011.03.052] [PMID] [PMCID]
Bonaglia, M. C., Giorda, R., Beri, S., De Agostini, C., Novara, F., & Fichera, M., et al. (2011). Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet, 7(7), e1002173.. [DOI:10.1371/journal.pgen.1002173] [PMID] [PMCID]
Bozdagi, O., Sakurai, T., Papapetrou, D., Wang, X., Dickstein, D. L., & Takahashi, N., et al. (2010). Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Molecular Autism, 1(1), 1-15. [DOI:10.1186/2040-2392-1-15] [PMID] [PMCID]
Durand, C. M., Betancur, C., Boeckers, T. M., Bockmann, J., Chaste, P., & Fauchereau, F., et al. (2007). Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genetics, 39(1), 25-7. [DOI:10.1038/ng1933] [PMID] [PMCID]
Durand, C. M., Perroy, J., Loll, F., Perrais, D., Fagni, L., & Bourgeron, T., et al. (2012). SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Molecular Psychiatry, 17(1), 71-84. [DOI:10.1038/mp.2011.57] [PMID] [PMCID]
Frenklach, A., Louie, S., Koop, M. M., & Bronte-Stewart, H. (2009). Excessive postural sway and the risk of falls at different stages of Parkinson's disease. Movement Disorders, 24(3), 377-85. [DOI:10.1002/mds.22358] [PMID]
Gauthier, J., Champagne, N., Lafrenière, R. G., Xiong, L., Spiegelman, D., & Brustein, E., et al. (2010). De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proceedings of the National Academy of Sciences, 107(17), 7863-8. [DOI:10.1073/pnas.0906232107] [PMID] [PMCID]
Gong, Y., Lippa, C. F., Zhu, J., Lin, Q., & Rosso, A. L. (2009). Disruption of glutamate receptors at Shank-postsynaptic platform in Alzheimer's disease. Brain Research, 1292, 191-8. [DOI:10.1016/j.brainres.2009.07.056] [PMID] [PMCID]
Jonsson, L., Zettergren, A., Pettersson, E., Hovey, D., Anckarsäter, H., & Westberg, L., et al. (2014). Association study between autistic-like traits and polymorphisms in the autism candidate regions RELN, CNTNAP2, SHANK3, and CDH9/10. Molecular Autism, 5(1), 1-9. [DOI:10.1186/2040-2392-5-55] [PMID] [PMCID]
Kreienkamp, H. J. (2008). Scaffolding proteins at the postsynaptic density: Shank as the architectural framework. Protein-Protein Interactions as New Drug Targets, 365-80. [DOI:10.1007/978-3-540-72843-6_15] [PMID]
Lepeta, K., Lourenco, M. V., Schweitzer, B. C., Martino Adami, P. V., Banerjee, P., & Catuara-Solarz, S., et al. (2016). Synaptopathies: synaptic dysfunction in neurological disorders–A review from students to students. Journal of neurochemistry, 138(6), 785-805. [DOI:10.1111/jnc.13713] [PMID] [PMCID]
Mameza, M. G., Dvoretskova, E., Bamann, M., Hönck, H. H., Güler, T., & Boeckers, T. M., et al. (2013). SHANK3 gene mutations associated with autism facilitate ligand binding to the Shank3 ankyrin repeat region. Journal of Biological Chemistry, 288(37), 26697-708. [DOI:10.1074/jbc.M112.424747] [PMID] [PMCID]
Mashayekhi, F., Mizban, N., Bidabadi, E., & Salehi, Z. (2016). The association of SHANK3 gene polymorphism and autism. Minerva Pediatrica. [Online Ahead of Print] [PMID]
Qin, J., Jia, M., Wang, L., Lu, T., Ruan, Y., & Liu, J., et al. (2009). Association study of SHANK3 gene polymorphisms with autism in Chinese Han population. BMC Medical Genetics, 10(1), 1-6. [DOI:10.1186/1471-2350-10-61] [PMID] [PMCID]
Qualmann, B., Boeckers, T. M., Jeromin, M., Gundelfinger, E. D., & Kessels, M. M. (2004). Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. Journal of Neuroscience, 24(10), 2481-95. [DOI:10.1523/JNEUROSCI.5479-03.2004] [PMID] [PMCID]
Rothermel, J. E., & Garcia, A. (1972). Treatment of hip fractures in patients with Parkinson's syndrome on levodopa therapy. Journal of Bone and Joint Surgery, 54(6), 1251-4. [DOI:10.2106/00004623-197254060-00013]
Roussignol, G., Ango, F., Romorini, S., Tu, J. C., Sala, C., & Worley, P. F., et al. (2005). Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. Journal of Neuroscience, 25(14), 3560-70. [DOI:10.1523/JNEUROSCI.4354-04.2005] [PMID] [PMCID]
Schapira, A. H. (1999) Science, medicine, and the future: Parkinson’s disease. The BMJ, 318, 311-4. [DOI:10.1136/bmj.318.7179.311] [PMID] [PMCID]
Shannak, K., Rajput, A., Rozdilsky, B., Kish, S., Gilbert, J., & Hornykiewicz, O. (1994). Noradrenaline, dopamine and serotonin levels and metabolism in the human hypothalamus: observations in Parkinson's disease and normal subjects. Brain Research, 639(1), 33-41. [DOI:10.1016/0006-8993(94)91761-2]
Shao, S., Xu, S., Yang, J., Zhang, T., He, Z., & Sun, Z., et al. (2014). A commonly carried genetic variant, rs9616915, in SHANK3 gene is associated with a reduced risk of autism spectrum disorder: Replication in a Chinese population. Molecular Biology Reports, 41(3), 1591-5. [DOI:10.1007/s11033-013-3005-5] [PMID]
Sykes, N. H., Toma, C., Wilson, N., Volpi, E. V., Sousa, I., & Pagnamenta, A. T., et al. (2009). Copy number variation and association analysis of SHANK3 as a candidate gene for autism in the IMGSAC collection. European Journal of Human Genetics, 17(10), 1347-53. [DOI:10.1038/ejhg.2009.47] [PMID] [PMCID]
Wilson, H. L., Wong, A. C. C., Shaw, S. R., Tse, W. Y., Stapleton, G. A., & Phelan, M. C., et al. (2003). Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. Journal of Medical Genetics, 40(8), 575-84. [DOI:10.1136/jmg.40.8.575] [PMID] [PMCID]
Received: 2018/01/6 | Accepted: 2018/04/30 | Published: 2021/01/1
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








